Genome-Wide Identification and Expression Analysis of nsLTP Gene Family in Rapeseed (Brassica napus) Reveals Their Critical Roles in Biotic and Abiotic Stress Responses
- PMID: 35955505
- PMCID: PMC9368849
- DOI: 10.3390/ijms23158372
Genome-Wide Identification and Expression Analysis of nsLTP Gene Family in Rapeseed (Brassica napus) Reveals Their Critical Roles in Biotic and Abiotic Stress Responses
Abstract
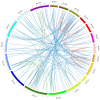
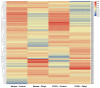
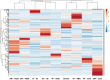
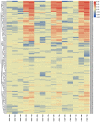
Non-specific lipid transfer proteins (nsLTPs) are small cysteine-rich basic proteins which play essential roles in plant growth, development and abiotic/biotic stress response. However, there is limited information about the nsLTP gene (BnLTP) family in rapeseed (Brassica napus). In this study, 283 BnLTP genes were identified in rapeseed, which were distributed randomly in 19 chromosomes of rapeseed. Phylogenetic analysis showed that BnLTP proteins were divided into seven groups. Exon/intron structure and MEME motifs both remained highly conserved in each BnLTP group. Segmental duplication and hybridization of rapeseed's two sub-genomes mainly contributed to the expansion of the BnLTP gene family. Various potential cis-elements that respond to plant growth, development, biotic/abiotic stresses, and phytohormone signals existed in BnLTP gene promoters. Transcriptome analysis showed that BnLTP genes were expressed in various tissues/organs with different levels and were also involved in the response to heat, drought, NaCl, cold, IAA and ABA stresses, as well as the treatment of fungal pathogens (Sclerotinia sclerotiorum and Leptosphaeria maculans). The qRT-PCR assay validated the results of RNA-seq expression analysis of two top Sclerotinia-responsive BnLTP genes, BnLTP129 and BnLTP161. Moreover, batches of BnLTPs might be regulated by BnTT1 and BnbZIP67 to play roles in the development, metabolism or adaptability of the seed coat and embryo in rapeseed. This work provides an important basis for further functional study of the BnLTP genes in rapeseed quality improvement and stress resistance.
Keywords: expression analysis; genome-wide identification; nsLTP; phylogenetic analysis; rapeseed (Brassica napus).
Conflict of interest statement
The authors declare that they have no known competing financial interest or personal relationships that could have appeared to influence the work reported in this paper.
Figures





Similar articles
-
Characterization of non-specific lipid transfer protein (nsLtp) gene families in the Brassica napus pangenome reveals abundance variation.BMC Plant Biol. 2022 Jan 7;22(1):21. doi: 10.1186/s12870-021-03408-5. BMC Plant Biol. 2022. PMID: 34996379 Free PMC article.
-
Brassica napus cytochrome P450 superfamily: Origin from parental species and involvement in diseases resistance, abiotic stresses tolerance, and seed quality traits.Ecotoxicol Environ Saf. 2024 Sep 15;283:116792. doi: 10.1016/j.ecoenv.2024.116792. Epub 2024 Aug 3. Ecotoxicol Environ Saf. 2024. PMID: 39096688
-
Genome-wide analysis and expression patterns of lipid phospholipid phospholipase gene family in Brassica napus L.BMC Genomics. 2021 Jul 18;22(1):548. doi: 10.1186/s12864-021-07862-1. BMC Genomics. 2021. PMID: 34273948 Free PMC article.
-
Systematic analysis of the non-specific lipid transfer protein gene family in Nicotiana tabacum reveal its potential roles in stress responses.Plant Physiol Biochem. 2022 Feb 1;172:33-47. doi: 10.1016/j.plaphy.2022.01.002. Epub 2022 Jan 5. Plant Physiol Biochem. 2022. PMID: 35016104 Review.
-
Phylogenomic analysis of 20S proteasome gene family reveals stress-responsive patterns in rapeseed (Brassica napus L.).Front Plant Sci. 2022 Oct 31;13:1037206. doi: 10.3389/fpls.2022.1037206. eCollection 2022. Front Plant Sci. 2022. PMID: 36388569 Free PMC article.
Cited by
-
Involvement of Pathogenesis-Related Proteins and Their Roles in Abiotic Stress Responses in Plants.Biomolecules. 2025 Jul 30;15(8):1103. doi: 10.3390/biom15081103. Biomolecules. 2025. PMID: 40867547 Free PMC article. Review.
-
Genome-wide identification of lipid transfer proteins in Sorghum bicolor and discovery of flower-specific promoters.Sci Rep. 2025 Jul 16;15(1):25710. doi: 10.1038/s41598-025-08625-8. Sci Rep. 2025. PMID: 40670441 Free PMC article.
-
Complex of Defense Polypeptides of Wheatgrass (Elytrigia elongata) Associated with Plant Immunity to Biotic and Abiotic Stress Factors.Plants (Basel). 2024 Sep 3;13(17):2459. doi: 10.3390/plants13172459. Plants (Basel). 2024. PMID: 39273943 Free PMC article.
-
The Small Key to the Treasure Chest: Endogenous Plant Peptides Involved in Symbiotic Interactions.Plants (Basel). 2025 Jan 26;14(3):378. doi: 10.3390/plants14030378. Plants (Basel). 2025. PMID: 39942939 Free PMC article. Review.
-
Systematic and functional analysis of non-specific lipid transfer protein family genes in sugarcane under Xanthomonas albilineans infection and salicylic acid treatment.Front Plant Sci. 2022 Oct 5;13:1014266. doi: 10.3389/fpls.2022.1014266. eCollection 2022. Front Plant Sci. 2022. PMID: 36275567 Free PMC article.
References
-
- Cui Y., Zeng X., Xiong Q., Wei D., Liao J., Xu Y., Chen G., Zhou Y., Dong H., Wan H., et al. Combining quantitative trait locus and co-expression analysis allowed identification of new candidates for oil accumulation in rapeseed. J. Exp. Bot. 2021;72:1649–1660. doi: 10.1093/jxb/eraa563. - DOI - PubMed
-
- Yan G., Yu P., Tian X., Guo L., Tu J., Shen J., Yi B., Fu T., Wen J., Liu K., et al. DELLA proteins BnaA6.RGA and BnaC7.RGA negatively regulate fatty acid biosynthesis by interacting with BnaLEC1s in Brassica napus. Plant Biotechnol. J. 2021;19:2011–2026. doi: 10.1111/pbi.13628. - DOI - PMC - PubMed
-
- Fitt B.D.L., Brun H., Barbetti M.J., Rimmer S.R. World-wide importance of phoma stem canker (Leptosphaeria maculans and L. biglobosa) on oilseed rape (Brassica napus) Eur. J. Plant Pathol. 2006;114:3–15. doi: 10.1007/s10658-005-2233-5. - DOI
MeSH terms
Substances
Grants and funding
- 32001441/National Natural Science Foundation of China
- 31871549/National Natural Science Foundation of China
- XDJK2020C038/Fundamental Research Funds for the Central Universities
- CY220219/Young Eagles Program of Chongqing Municipal Commission of Education
- S202110635162/Chongqing Municipal Training Program of Innovation and Entrepreneurship for Undergraduates
LinkOut - more resources
Full Text Sources

