Aggregated Genomic Data as Cohort-Specific Allelic Frequencies can Boost Variants and Genes Prioritization in Non-Solved Cases of Inherited Retinal Dystrophies
- PMID: 35955564
- PMCID: PMC9368980
- DOI: 10.3390/ijms23158431
Aggregated Genomic Data as Cohort-Specific Allelic Frequencies can Boost Variants and Genes Prioritization in Non-Solved Cases of Inherited Retinal Dystrophies
Abstract
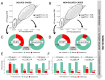
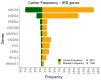
The introduction of NGS in genetic diagnosis has increased the repertoire of variants and genes involved and the amount of genomic information produced. We built an allelic-frequency (AF) database for a heterogeneous cohort of genetic diseases to explore the aggregated genomic information and boost diagnosis in inherited retinal dystrophies (IRD). We retrospectively selected 5683 index-cases with clinical exome sequencing tests available, 1766 with IRD and the rest with diverse genetic diseases. We calculated a subcohort's IRD-specific AF and compared it with suitable pseudocontrols. For non-solved IRD cases, we prioritized variants with a significant increment of frequencies, with eight variants that may help to explain the phenotype, and 10/11 of uncertain significance that were reclassified as probably pathogenic according to ACMG. Moreover, we developed a method to highlight genes with more frequent pathogenic variants in IRD cases than in pseudocontrols weighted by the increment of benign variants in the same comparison. We identified 18 genes for further studies that provided new insights in five cases. This resource can also help one to calculate the carrier frequency in IRD genes. A cohort-specific AF database assists with variants and genes prioritization and operates as an engine that provides a new hypothesis in non-solved cases, augmenting the diagnosis rate.
Keywords: carrier frequency; gene prioritization; genetic rare diseases; inherited retinal dystrophies; variant prioritization; variants of uncertain significance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
[Comparison study of whole exome sequencing and targeted panel sequencing in molecular diagnosis of inherited retinal dystrophies].Beijing Da Xue Xue Bao Yi Xue Ban. 2020 Oct 18;52(5):836-844. doi: 10.19723/j.issn.1671-167X.2020.05.007. Beijing Da Xue Xue Bao Yi Xue Ban. 2020. PMID: 33047716 Free PMC article. Chinese.
-
Novel Candidate Genes and a Wide Spectrum of Structural and Point Mutations Responsible for Inherited Retinal Dystrophies Revealed by Exome Sequencing.PLoS One. 2016 Dec 22;11(12):e0168966. doi: 10.1371/journal.pone.0168966. eCollection 2016. PLoS One. 2016. PMID: 28005958 Free PMC article.
-
Genetic spectrum of retinal dystrophies in Tunisia.Sci Rep. 2020 Jul 8;10(1):11199. doi: 10.1038/s41598-020-67792-y. Sci Rep. 2020. PMID: 32641690 Free PMC article.
-
Scaling New Heights in the Genetic Diagnosis of Inherited Retinal Dystrophies.Adv Exp Med Biol. 2019;1185:215-219. doi: 10.1007/978-3-030-27378-1_35. Adv Exp Med Biol. 2019. PMID: 31884614 Review.
-
Clinical and genetic spectrums of 413 North African families with inherited retinal dystrophies and optic neuropathies.Orphanet J Rare Dis. 2022 May 12;17(1):197. doi: 10.1186/s13023-022-02340-7. Orphanet J Rare Dis. 2022. PMID: 35551639 Free PMC article. Review.
Cited by
-
Five years' experience of the clinical exome sequencing in a Spanish single center.Sci Rep. 2022 Nov 10;12(1):19209. doi: 10.1038/s41598-022-23786-6. Sci Rep. 2022. PMID: 36357507 Free PMC article.
-
Identification of new families and variants in autosomal dominant macular dystrophy associated with THRB.Sci Rep. 2025 Apr 28;15(1):14904. doi: 10.1038/s41598-025-97768-9. Sci Rep. 2025. PMID: 40295579 Free PMC article.
-
Medical Genetics, Genomics and Bioinformatics-2022.Int J Mol Sci. 2023 May 18;24(10):8968. doi: 10.3390/ijms24108968. Int J Mol Sci. 2023. PMID: 37240312 Free PMC article.
-
Prioritization of New Candidate Genes for Rare Genetic Diseases by a Disease-Aware Evaluation of Heterogeneous Molecular Networks.Int J Mol Sci. 2023 Jan 14;24(2):1661. doi: 10.3390/ijms24021661. Int J Mol Sci. 2023. PMID: 36675175 Free PMC article.
References
-
- Sanchez-Navarro I., Da Silva L.R.J.R.J., Blanco-Kelly F., Zurita O., Sanchez-Bolivar N., Villaverde C., Lopez-Molina M.I.I., Garcia-Sandoval B., Tahsin-Swafiri S., Minguez P., et al. Combining targeted panel-based resequencing and copy-number variation analysis for the diagnosis of inherited syndromic retinopathies and associated ciliopathies. Sci. Rep. 2018;8:5285. doi: 10.1038/s41598-018-23520-1. - DOI - PMC - PubMed
MeSH terms
Grants and funding
- B2017/BMD-3721/Comunidad de Madrid
- PI18/00579/Instituto de Salud Carlos III
- PI19/00321/Instituto de Salud Carlos III
- PI20/00851/Instituto de Salud Carlos III
- 4019/012/Fundación Ramón Areces
- 06/07/0036/Centre for Biomedical Network Research on Rare Diseases
- PT13/0010/0012/IIS-FJD Biobank
- ONCE/Organización Nacional de Ciegos Españoles
- PEJ-2017-AI/BMD7256/Comunidad de Madrid
- IMP/00019/Instituto de Salud Carlos III
- FI17/00192/Instituto de Salud Carlos III
- PEJ-2020-AI/BMD-18610/Comunidad de Madrid
- CA18/00017/Instituto de Salud Carlos III
- 2019-T2/BMD-13714/Comunidad de Madrid
- JR17/00020/Instituto de Salud Carlos III
- FI18/00123/Instituto de Salud Carlos III
- CP16/00116, CPII21/00015/Instituto de Salud Carlos III
LinkOut - more resources
Full Text Sources

