Tailoring the Structure of Cell Penetrating DNA and RNA Binding Nucleopeptides
- PMID: 35955638
- PMCID: PMC9369335
- DOI: 10.3390/ijms23158504
Tailoring the Structure of Cell Penetrating DNA and RNA Binding Nucleopeptides
Abstract
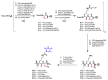
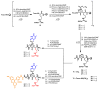
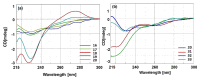
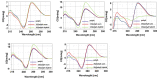
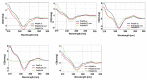
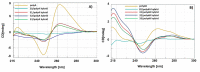
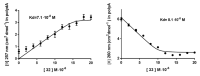
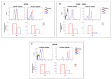
Synthetic nucleic acid interactors represent an exciting research field due to their biotechnological and potential therapeutic applications. The translation of these molecules into drugs is a long and difficult process that justifies the continuous research of new chemotypes endowed with favorable binding, pharmacokinetic and pharmacodynamic properties. In this scenario, we describe the synthesis of two sets of homo-thymine nucleopeptides, in which nucleobases are inserted in a peptide structure, to investigate the role of the underivatized amino acid residue and the distance of the nucleobase from the peptide backbone on the nucleic acid recognition process. It is worth noting that the CD spectroscopy investigation showed that two of the reported nucleopeptides, consisting of alternation of thymine functionalized L-Orn and L-Dab and L-Arg as underivatized amino acids, were able to efficiently bind DNA and RNA targets and cross both cell and nuclear membranes.
Keywords: cell penetrating peptide synthesis; cell penetrating peptides; nuclear localization sequences; nucleic acid binders; nucleopeptides.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Belmont P., Constant J.F., Demeunynck M. Nucleic acid conformation diversity: From structure to function and regulation. Chem. Soc. Rev. 2001;30:70–81. doi: 10.1039/a904630e. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

