Transcriptional and Post-Translational Regulation of Junctional Adhesion Molecule-B (JAM-B) in Leukocytes under Inflammatory Stimuli
- PMID: 35955781
- PMCID: PMC9369439
- DOI: 10.3390/ijms23158646
Transcriptional and Post-Translational Regulation of Junctional Adhesion Molecule-B (JAM-B) in Leukocytes under Inflammatory Stimuli
Abstract
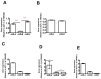
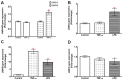
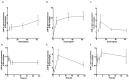
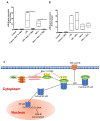
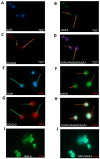
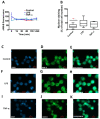
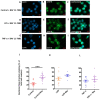
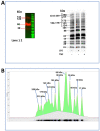
Junctional adhesion molecules (JAMs; comprising JAM-A, -B and -C) act as receptors for viruses, mediate cell permeability, facilitate leukocyte migration during sterile and non-sterile inflammation and are important for the maintenance of epithelial barrier integrity. As such, they are implicated in the development of both communicable and non-communicable chronic diseases. Here, we investigated the expression and regulation of JAM-B in leukocytes under pathogen- and host-derived inflammatory stimuli using immunoassays, qPCR and pharmacological inhibitors of inflammatory signalling pathways. We show that JAM-B is expressed at both the mRNA and protein level in leukocytes. JAM-B protein is localised to the cytoplasm, Golgi apparatus and in the nucleus around ring-shaped structures. We also provide evidence that JAM-B nuclear localisation occurs via the classical importin-α/β pathway, which is likely mediated through JAM-B protein nuclear localisation signals (NLS) and export signals (NES). In addition, we provide evidence that under both pathogen- and host-derived inflammatory stimuli, JAM-B transcription is regulated via the NF-κB-dependent pathways, whereas at the post-translational level JAM-B is regulated by ubiquitin-proteosome pathways. Anaphase-promoting ubiquitin ligase complex (APC/C) and herpes simplex virus-associated ubiquitin-specific protease (HAUSP/USP) were identified as candidates for JAM-B ubiquitination and de-ubiquitination, respectively. The expression and regulation of JAM-B in leukocytes reported here is a novel observation and contrasts with previous reports. The data reported here suggest that JAM-B expression in leukocytes is under the control of common inflammatory pathways.
Keywords: barrier function; cell adhesion; cell migration; cell permeability; host; inflammation; pathogen; tight junctions.
Conflict of interest statement
None of the authors have declared a conflict of interest.
Figures



References
-
- Li X., Stankovic M., Lee B.P.-L., Aurrand-Lions M., Hahn C.N., Lu Y., Imhof B.A., Vadas M.A., Gamble J.R. JAM-C induces endothelial cell permeability through its association and regulation of β3 integrins. Arterioscler. Thromb. Vasc. Biol. 2009;29:1200–1206. doi: 10.1161/ATVBAHA.109.189217. - DOI - PubMed
MeSH terms
Substances
Grants and funding
- BB/R012512/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/F/000PR10343/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/F/000PR10347/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- PG/16/94/32393/BHF_/British Heart Foundation/United Kingdom
LinkOut - more resources
Full Text Sources
Research Materials

