Structural Evolution of Delta (B.1.617.2) and Omicron (BA.1) Spike Glycoproteins
- PMID: 35955815
- PMCID: PMC9369368
- DOI: 10.3390/ijms23158680
Structural Evolution of Delta (B.1.617.2) and Omicron (BA.1) Spike Glycoproteins
Abstract
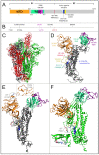
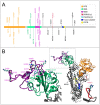
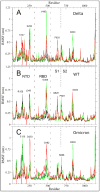
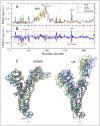
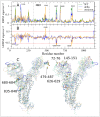
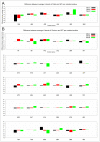
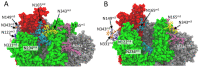
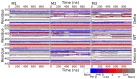
The vast amount of epidemiologic and genomic data that were gathered as a global response to the COVID-19 pandemic that was caused by SARS-CoV-2 offer a unique opportunity to shed light on the structural evolution of coronaviruses and in particular on the spike (S) glycoprotein, which mediates virus entry into the host cell by binding to the human ACE2 receptor. Herein, we carry out an investigation into the dynamic properties of the S glycoprotein, focusing on the much more transmissible Delta and Omicron variants. Notwithstanding the great number of mutations that have accumulated, particularly in the Omicron S glycoprotein, our data clearly showed the conservation of some structural and dynamic elements, such as the global motion of the receptor binding domain (RBD). However, our studies also revealed structural and dynamic alterations that were concentrated in the aa 627-635 region, on a small region of the receptor binding motif (aa 483-485), and the so-called "fusion-peptide proximal region". In particular, these last two S regions are known to be involved in the human receptor ACE2 recognition and membrane fusion. Our structural evidence, therefore, is likely involved in the observed different transmissibility of these S mutants. Finally, we highlighted the role of glycans in the increased RBD flexibility of the monomer in the up conformation of Omicron.
Keywords: COVID-19; SARS-CoV-2; delta; molecular dynamics; omicron; spike; variants.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- World Health Organization. [(accessed on 26 May 2022)]. Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants.
-
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., et al. The Species Severe Acute Respiratory Syndrome-Related Coronavirus: Classifying 2019-NCoV and Naming It SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
- H2020-SC1-PHE-CORONAVIRUS-2020/EU
- Departments of Excellence-2018' Program (Dipartimenti di Eccellenza) to DIBAF-Department of University of Tuscia, Project 'Landscape 4.0-food, wellbeing and environment'./Ministry of Education, Universities and Research
- Project 5M-2020-23682104/Ministero della Salute, 5 per Mille
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

