c-Kit Induces Migration of Triple-Negative Breast Cancer Cells and Is a Promising Target for Tyrosine Kinase Inhibitor Treatment
- PMID: 35955836
- PMCID: PMC9369219
- DOI: 10.3390/ijms23158702
c-Kit Induces Migration of Triple-Negative Breast Cancer Cells and Is a Promising Target for Tyrosine Kinase Inhibitor Treatment
Abstract
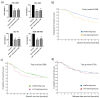
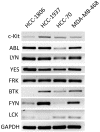
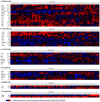
Triple-negative breast cancer (TNBC) is associated with a poor prognosis and the absence of targeted therapy. c-Kit, a receptor tyrosine kinase (RTK), is considered a molecular target for anticancer drugs. Tyrosine kinase inhibitors (TKIs) recognizing c-Kit are used for the treatment of c-Kit-expressing tumors. However, the expression, function, and therapeutic potential of c-Kit have been little explored in TNBC. Here, we studied the expression and effects of c-Kit in TNBC through in vitro and in silico analysis, and evaluated the response to TKIs targeting c-Kit. Analysis of TNBC cells showed the expression of functional c-Kit at the cell membrane. The stimulation of c-Kit with its ligand induced the activation of STAT3, Akt, and ERK1/2, increasing cell migration, but had no effect on cell proliferation or response to Doxorubicin. Analysis of public datasets showed that the expression of c-Kit in tumors was not associated with patient survival. Finally, TNBC cells were susceptible to TKIs, in particular the effect of Nilotinib was stronger than Doxorubicin in all cell lines. In conclusion, TNBC cells express functional c-Kit, which is a targetable molecule, and show a strong response to Nilotinib that may be considered a candidate drug for the treatment of TNBC.
Keywords: c-Kit; nilotinib; receptor tyrosine kinase; stem cell factor; triple-negative breast cancer; tyrosine kinase inhibitor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Lehmann B.D., Jovanovic B., Chen X., Estrada M.V., Johnson K.N., Shyr Y., Moses H.L., Sanders M.E., Pietenpol J.A. Refinement of triple-negative breast cancer molecular subtypes: Implications for neoadyuvant chemotherapy selection. PLoS ONE. 2016;11:e0157368. doi: 10.1371/journal.pone.0157368. - DOI - PMC - PubMed
-
- Burstein M.D., Tsimelzon A., Poage G.M., Covington K.R., Contreras A., Fuqua S.A.W., Savage M.I., Osborne C.K., Hilsenbeck S.G., Chang J.C., et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin. Cancer Res. 2015;21:1688–1698. doi: 10.1158/1078-0432.CCR-14-0432. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

