Unifying Different Cancer Theories in a Unique Tumour Model: Chronic Inflammation and Deaminases as Meeting Points
- PMID: 35955853
- PMCID: PMC9368936
- DOI: 10.3390/ijms23158720
Unifying Different Cancer Theories in a Unique Tumour Model: Chronic Inflammation and Deaminases as Meeting Points
Abstract
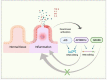
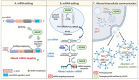
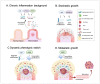
The increase in cancer incidences shows that there is a need to better understand tumour heterogeneity to achieve efficient treatments. Interestingly, there are several common features among almost all types of cancers, with chronic inflammation induction and deaminase dysfunctions singled out. Deaminases are a family of enzymes with nucleotide-editing capacity, which are classified into two main groups: DNA-based and RNA-based. Remarkably, a close relationship between inflammation and the dysregulation of these molecules has been widely documented, which may explain the characteristic intratumor heterogeneity, both at DNA and transcriptional levels. Indeed, heterogeneity in cancer makes it difficult to establish a unique tumour progression model. Currently, there are three main cancer models-stochastic, hierarchic, and dynamic-although there is no consensus on which one better resembles cancer biology because they are usually overly simplified. Here, to accurately explain tumour progression, we propose interactions among chronic inflammation, deaminases dysregulation, intratumor genetic heterogeneity, cancer phenotypic plasticity, and even the previously proposed appearance of cancer stem-like cell populations in the edges of advanced solid tumour masses (instead of being the cells of origin of primary malignancies). The new tumour development model proposed in this study does not contradict previously accepted models and it may open up a window to interesting therapeutic approaches.
Keywords: ADAR; AID; APOBEC; cancer phenotype plasticity; cancer stem cells; deaminases dysregulation; tumour development model.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
- SOMM17/6109/UGR/Consejería de Economía, Conocimiento, Empresas y Universidad de la Junta de Andalucía
- BIO-349/University of Jaen, Acción I apoyo a la investigación
- PEMP-0205-2020/Consejería de Salud y Familias de la Junta de Andalucía
- PIE16/00045/Ministry of Economy and Competitiveness
- RTI2018-101309-B-C22/Ministry of Science, Innovation and Universities

