Reference Genes across Nine Brain Areas of Wild Type and Prader-Willi Syndrome Mice: Assessing Differences in Igfbp7, Pcsk1, Nhlh2 and Nlgn3 Expression
- PMID: 35955861
- PMCID: PMC9369261
- DOI: 10.3390/ijms23158729
Reference Genes across Nine Brain Areas of Wild Type and Prader-Willi Syndrome Mice: Assessing Differences in Igfbp7, Pcsk1, Nhlh2 and Nlgn3 Expression
Abstract
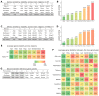
Prader−Willi syndrome (PWS) is a complex neurodevelopmental disorder caused by the deletion or inactivation of paternally expressed imprinted genes at the chromosomal region 15q11−q13. The PWS-critical region (PWScr) harbors tandemly repeated non-protein coding IPW-A exons hosting the intronic SNORD116 snoRNA gene array that is predominantly expressed in brain. Paternal deletion of PWScr is associated with key PWS symptoms in humans and growth retardation in mice (PWScr model). Dysregulation of the hypothalamic−pituitary axis (HPA) is thought to be causally involved in the PWS phenotype. Here we performed a comprehensive reverse transcription quantitative PCR (RT-qPCR) analysis across nine different brain regions of wild-type (WT) and PWScr mice to identify stably expressed reference genes. Four methods (Delta Ct, BestKeeper, Normfinder and Genorm) were applied to rank 11 selected reference gene candidates according to their expression stability. The resulting panel consists of the top three most stably expressed genes suitable for gene-expression profiling and comparative transcriptome analysis of WT and/or PWScr mouse brain regions. Using these reference genes, we revealed significant differences in the expression patterns of Igfbp7, Nlgn3 and three HPA associated genes: Pcsk1, Pcsk2 and Nhlh2 across investigated brain regions of wild-type and PWScr mice. Our results raise a reasonable doubt on the involvement of the Snord116 in posttranscriptional regulation of Nlgn3 and Nhlh2 genes. We provide a valuable tool for expression analysis of specific genes across different areas of the mouse brain and for comparative investigation of PWScr mouse models to discover and verify different regulatory pathways affecting this complex disorder.
Keywords: Igfbp7; Nhlh2; Nlgn3; PWS-critical region; Pcsk1; Pcsk2; Prader–Willi syndrome; RT-qPCR; SNORD116; brain; brain regions; gene expression; posttranscriptional regulation; reference genes; transcriptome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Maternal transcription of non-protein coding RNAs from the PWS-critical region rescues growth retardation in mice.Sci Rep. 2016 Feb 5;6:20398. doi: 10.1038/srep20398. Sci Rep. 2016. PMID: 26848093 Free PMC article.
-
Deletion of the Snord116/SNORD116 Alters Sleep in Mice and Patients with Prader-Willi Syndrome.Sleep. 2016 Mar 1;39(3):637-44. doi: 10.5665/sleep.5542. Sleep. 2016. PMID: 26446116 Free PMC article.
-
Deficiency in prohormone convertase PC1 impairs prohormone processing in Prader-Willi syndrome.J Clin Invest. 2017 Jan 3;127(1):293-305. doi: 10.1172/JCI88648. Epub 2016 Dec 12. J Clin Invest. 2017. PMID: 27941249 Free PMC article.
-
A Comprehensive Review of Genetically Engineered Mouse Models for Prader-Willi Syndrome Research.Int J Mol Sci. 2021 Mar 31;22(7):3613. doi: 10.3390/ijms22073613. Int J Mol Sci. 2021. PMID: 33807162 Free PMC article. Review.
-
Cognitive deficits in the Snord116 deletion mouse model for Prader-Willi syndrome.Neurobiol Learn Mem. 2019 Nov;165:106874. doi: 10.1016/j.nlm.2018.05.011. Epub 2018 May 23. Neurobiol Learn Mem. 2019. PMID: 29800646 Free PMC article. Review.
Cited by
-
Evaluation of suitable reference genes for gene expression studies in the developing mouse cortex using RT-qPCR.BMC Neurosci. 2025 Feb 18;26(1):12. doi: 10.1186/s12868-025-00934-y. BMC Neurosci. 2025. PMID: 39966711 Free PMC article.
-
Identification of Reliable Reference Genes for Use in Gene Expression Studies in Rat Febrile Seizure Model.Int J Mol Sci. 2024 Oct 16;25(20):11125. doi: 10.3390/ijms252011125. Int J Mol Sci. 2024. PMID: 39456907 Free PMC article.
-
Footprints in the Sno: investigating the cellular and molecular mechanisms of SNORD116.Open Biol. 2025 Mar;15(3):240371. doi: 10.1098/rsob.240371. Epub 2025 Mar 19. Open Biol. 2025. PMID: 40101781 Free PMC article. Review.
-
Insulin secretion deficits in a Prader-Willi syndrome β-cell model are associated with a concerted downregulation of multiple endoplasmic reticulum chaperones.PLoS Genet. 2023 Apr 17;19(4):e1010710. doi: 10.1371/journal.pgen.1010710. eCollection 2023 Apr. PLoS Genet. 2023. PMID: 37068109 Free PMC article.
References
-
- Cavaillé J., Buiting K., Kiefmann M., Lalande M., Brannan C.I., Horsthemke B., Bachellerie J.-P., Brosius J., Hüttenhofer A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl. Acad. Sci. USA. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

