Exploring Ligand Binding Domain Dynamics in the NRs Superfamily
- PMID: 35955864
- PMCID: PMC9369052
- DOI: 10.3390/ijms23158732
Exploring Ligand Binding Domain Dynamics in the NRs Superfamily
Abstract
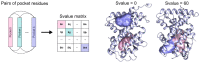
Nuclear receptors (NRs) are transcription factors that play an important role in multiple diseases, such as cancer, inflammation, and metabolic disorders. They share a common structural organization composed of five domains, of which the ligand-binding domain (LBD) can adopt different conformations in response to substrate, agonist, and antagonist binding, leading to distinct transcription effects. A key feature of NRs is, indeed, their intrinsic dynamics that make them a challenging target in drug discovery. This work aims to provide a meaningful investigation of NR structural variability to outline a dynamic profile for each of them. To do that, we propose a methodology based on the computation and comparison of protein cavities among the crystallographic structures of NR LBDs. First, pockets were detected with the FLAPsite algorithm and then an "all against all" approach was applied by comparing each pair of pockets within the same sub-family on the basis of their similarity score. The analysis concerned all the detectable cavities in NRs, with particular attention paid to the active site pockets. This approach can guide the investigation of NR intrinsic dynamics, the selection of reference structures to be used in drug design and the easy identification of alternative binding sites.
Keywords: agonists; antagonists; drug design; flexibility; ligand binding domain; nuclear receptors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Structural overview and perspectives of the nuclear receptors, a major family as the direct targets for small-molecule drugs.Acta Biochim Biophys Sin (Shanghai). 2022 Jan 25;54(1):12-24. doi: 10.3724/abbs.2021001. Acta Biochim Biophys Sin (Shanghai). 2022. PMID: 35130630 Free PMC article.
-
Covalent ligands of nuclear receptors.Eur J Med Chem. 2023 Dec 5;261:115869. doi: 10.1016/j.ejmech.2023.115869. Epub 2023 Oct 11. Eur J Med Chem. 2023. PMID: 37857142 Review.
-
Conformational adaptation of nuclear receptor ligand binding domains to agonists: potential for novel approaches to ligand design.J Steroid Biochem Mol Biol. 2005 Feb;93(2-5):127-37. doi: 10.1016/j.jsbmb.2005.01.004. Epub 2005 Feb 19. J Steroid Biochem Mol Biol. 2005. PMID: 15860255 Review.
-
Non-canonical modulators of nuclear receptors.Bioorg Med Chem Lett. 2016 Sep 1;26(17):4157-64. doi: 10.1016/j.bmcl.2016.07.067. Epub 2016 Jul 29. Bioorg Med Chem Lett. 2016. PMID: 27503683 Review.
-
Exploring Flexibility of Progesterone Receptor Ligand Binding Domain Using Molecular Dynamics.PLoS One. 2016 Nov 8;11(11):e0165824. doi: 10.1371/journal.pone.0165824. eCollection 2016. PLoS One. 2016. PMID: 27824891 Free PMC article.
Cited by
-
Progress of the "Molecular Informatics" Section in 2022.Int J Mol Sci. 2023 May 29;24(11):9442. doi: 10.3390/ijms24119442. Int J Mol Sci. 2023. PMID: 37298393 Free PMC article.
-
Liver X Receptors (LXRs) in cancer-an Eagle's view on molecular insights and therapeutic opportunities.Front Cell Dev Biol. 2024 Mar 14;12:1386102. doi: 10.3389/fcell.2024.1386102. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38550382 Free PMC article. Review.
-
Nuclear receptors in health and disease: signaling pathways, biological functions and pharmaceutical interventions.Signal Transduct Target Ther. 2025 Jul 28;10(1):228. doi: 10.1038/s41392-025-02270-3. Signal Transduct Target Ther. 2025. PMID: 40717128 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

