diaPASEF Proteomics and Feature Selection for the Description of Sputum Proteome Profiles in a Cohort of Different Subtypes of Lung Cancer Patients and Controls
- PMID: 35955870
- PMCID: PMC9369298
- DOI: 10.3390/ijms23158737
diaPASEF Proteomics and Feature Selection for the Description of Sputum Proteome Profiles in a Cohort of Different Subtypes of Lung Cancer Patients and Controls
Abstract
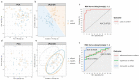
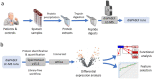
The high mortality, the presence of an initial asymptomatic stage and the fact that diagnosis in early stages reduces mortality justify the implementation of screening programs in the populations at risk of lung cancer. It is imperative to develop less aggressive methods that can complement existing diagnosis technologies. In this study, we aimed to identify lung cancer protein biomarkers and pathways affected in sputum samples, using the recently developed diaPASEF mass spectrometry (MS) acquisition mode. The sputum proteome of lung cancer cases and controls was analyzed through nano-HPLC-MS using the diaPASEF mode. For functional analysis, the results from differential expression analysis were further analyzed in the STRING platform, and feature selection was performed using sparse partial least squares discriminant analysis (sPLS-DA). Our results showed an activation of inflammation, with an alteration of pathways and processes related to acute-phase, complement, and immune responses. The resulting sPLS-DA model separated between case and control groups with high levels of sensitivity and specificity. In conclusion, we showed how new-generation proteomics can be used to detect potential biomarkers in sputum samples, and ultimately to discriminate patients from controls and even to help to differentiate between different cancer subtypes.
Keywords: adenocarcinoma; diaPASEF; lung cancer; proteomics; sputum.
Conflict of interest statement
All authors declare that there were no financial/commercial conflict of interest in this study.
Figures



Similar articles
-
Proteome screening of pleural effusions identifies galectin 1 as a diagnostic biomarker and highlights several prognostic biomarkers for malignant mesothelioma.Mol Cell Proteomics. 2014 Mar;13(3):701-15. doi: 10.1074/mcp.M113.030775. Epub 2013 Dec 20. Mol Cell Proteomics. 2014. PMID: 24361865 Free PMC article.
-
Identification of ENO1 as a potential sputum biomarker for early-stage lung cancer by shotgun proteomics.Clin Lung Cancer. 2014 Sep;15(5):372-378.e1. doi: 10.1016/j.cllc.2014.05.003. Epub 2014 Jun 3. Clin Lung Cancer. 2014. PMID: 24984566 Free PMC article.
-
Proteome analysis of non-small cell lung cancer cell line secretomes and patient sputum reveals biofluid biomarker candidates for cisplatin response prediction.J Proteomics. 2019 Mar 30;196:106-119. doi: 10.1016/j.jprot.2019.01.018. Epub 2019 Jan 30. J Proteomics. 2019. PMID: 30710758 Clinical Trial.
-
Proteomic Analysis of Human Sputum for the Diagnosis of Lung Disorders: Where Are We Today?Int J Mol Sci. 2022 May 19;23(10):5692. doi: 10.3390/ijms23105692. Int J Mol Sci. 2022. PMID: 35628501 Free PMC article. Review.
-
Molecular sputum analysis for the diagnosis of lung cancer.Br J Cancer. 2013 Aug 6;109(3):530-7. doi: 10.1038/bjc.2013.393. Epub 2013 Jul 18. Br J Cancer. 2013. PMID: 23868001 Free PMC article. Review.
Cited by
-
Towards Precision Prognostication and Personalized Therapeutics through Proteomics.Int J Mol Sci. 2023 Mar 28;24(7):6361. doi: 10.3390/ijms24076361. Int J Mol Sci. 2023. PMID: 37047334 Free PMC article.
-
Alpha-1 Antitrypsin as a Regulatory Protease Inhibitor Modulating Inflammation and Shaping the Tumor Microenvironment in Cancer.Cells. 2025 Jan 10;14(2):88. doi: 10.3390/cells14020088. Cells. 2025. PMID: 39851516 Free PMC article. Review.
References
-
- The National Lung Screening Trial Research Team. Aberle D.R., Adams A.M., Berg C.D., Black W.C., Clapp J.D., Fagerstrom R.M., Gareen I.F., Gatsonis C., Marcus P.M., et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N. Engl. J. Med. 2011;365:395–409. doi: 10.1056/NEJMoa1102873. - DOI - PMC - PubMed

