Combined QTL Mapping across Multiple Environments and Co-Expression Network Analysis Identified Key Genes for Embryogenic Callus Induction from Immature Maize Embryos
- PMID: 35955919
- PMCID: PMC9368897
- DOI: 10.3390/ijms23158786
Combined QTL Mapping across Multiple Environments and Co-Expression Network Analysis Identified Key Genes for Embryogenic Callus Induction from Immature Maize Embryos
Abstract
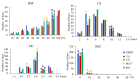
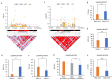
The ability of immature embryos to induce embryogenic callus (EC) is crucial for genetic transformation in maize, which is highly genotype-dependent. To dissect the genetic basis of maize EC induction, we conducted QTL mapping for four EC induction-related traits, the rate of embryogenic callus induction (REC), rate of shoot formation (RSF), length of shoot (LS), and diameter of callus (DC) under three environments by using an IBM Syn10 DH population derived from a cross of B73 and Mo17. These EC induction traits showed high broad-sense heritability (>80%), and significantly negative correlations were observed between REC and each of the other traits across multiple environments. A total of 41 QTLs for EC induction were identified, among which 13, 12, 10, and 6 QTLs were responsible for DC, RSF, LS, and REC, respectively. Among them, three major QTLs accounted for >10% of the phenotypic variation, including qLS1-1 (11.54%), qLS1-3 (10.68%), and qREC4-1 (11.45%). Based on the expression data of the 215 candidate genes located in these QTL intervals, we performed a weighted gene co-expression network analysis (WGCNA). A combined use of KEGG pathway enrichment and eigengene-based connectivity (KME) values identified the EC induction-associated module and four hub genes (Zm00001d028477, Zm00001d047896, Zm00001d034388, and Zm00001d022542). Gene-based association analyses validated that the variations in Zm00001d028477 and Zm00001d034388, which were involved in tryptophan biosynthesis and metabolism, respectively, significantly affected EC induction ability among different inbred lines. Our study brings novel insights into the genetic and molecular mechanisms of EC induction and helps to promote marker-assisted selection of high-REC varieties in maize.
Keywords: QTL mapping; WGCNA; candidate gene; embryogenic callus; immature embryo; maize.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Satish L., Rathinapriya P., Ceasar S.A., Rency A.S., Pandian S., Rameshkumar R., Subramanian A., Ramesh M. Effects of cefotaxime, amino acids and carbon source on somatic embryogenesis and plant regeneration in four Indian genotypes of foxtail millet (Setaria italica L.) In Vitr. Cell. Dev. Biol. Plant. 2015;52:140–153. doi: 10.1007/s11627-015-9724-7. - DOI
-
- Saha S., Islam Z., Islam S., Hassan M.F., Hossain M.S., Islam S.M.S. Enhancement of Somatic Embryogenesis by Mature and Immature Seeds in Wheat (Triticum aestivum L.) J. Biol. Life Sci. 2017;8:20. doi: 10.5296/jbls.v8i2.11529. - DOI
-
- Ana L.M.L., Ivone B.d.O.E., Carlos A.S., Claudete A.M., Maria d.F.P.S.M. Somatic embryogenesis and plant regeneration in popcorn (Zea mays L.) inbred lines. Afr. J. Biotechnol. 2017;16:1738–1742. doi: 10.5897/AJB2016.15767. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

