Computational Analysis of Short Linear Motifs in the Spike Protein of SARS-CoV-2 Variants Provides Possible Clues into the Immune Hijack and Evasion Mechanisms of Omicron Variant
- PMID: 35955954
- PMCID: PMC9368778
- DOI: 10.3390/ijms23158822
Computational Analysis of Short Linear Motifs in the Spike Protein of SARS-CoV-2 Variants Provides Possible Clues into the Immune Hijack and Evasion Mechanisms of Omicron Variant
Abstract
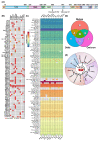
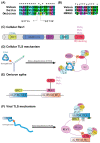
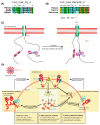
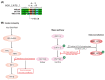
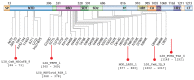
Short linear motifs (SLiMs) are short linear sequences that can mediate protein-protein interaction. Mimicking eukaryotic SLiMs to compete with extra- or intracellular binding partners, or to sequester host proteins is the crucial strategy of viruses to pervert the host system. Evolved proteins in viruses facilitate minimal protein-protein interactions that significantly affect intracellular signaling networks. Unfortunately, very little information about SARS-CoV-2 SLiMs is known, especially across SARS-CoV-2 variants. Through the ELM database-based sequence analysis of spike proteins from all the major SARS-CoV-2 variants, we identified four overriding SLiMs in the SARS-CoV-2 Omicron variant, namely, LIG_TRFH_1, LIG_REV1ctd_RIR_1, LIG_CaM_NSCaTE_8, and MOD_LATS_1. These SLiMs are highly likely to interfere with various immune functions, interact with host intracellular proteins, regulate cellular pathways, and lubricate viral infection and transmission. These cellular interactions possibly serve as potential therapeutic targets for these variants, and this approach can be further exploited to combat emerging SARS-CoV-2 variants.
Keywords: COVID-19; Omicron; SARS-CoV-2; SLiMs; coronaviruses; motifs; spike protein; variant.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Uncovering potential host proteins and pathways that may interact with eukaryotic short linear motifs in viral proteins of MERS, SARS and SARS2 coronaviruses that infect humans.PLoS One. 2021 Feb 3;16(2):e0246150. doi: 10.1371/journal.pone.0246150. eCollection 2021. PLoS One. 2021. PMID: 33534852 Free PMC article.
-
Transformations, Lineage Comparisons, and Analysis of Down-to-Up Protomer States of Variants of the SARS-CoV-2 Prefusion Spike Protein, Including the UK Variant B.1.1.7.Microbiol Spectr. 2021 Sep 3;9(1):e0003021. doi: 10.1128/Spectrum.00030-21. Epub 2021 Aug 4. Microbiol Spectr. 2021. PMID: 34346753 Free PMC article.
-
Characterization of the novel SARS-CoV-2 Omicron (B.1.1.529) variant of concern and its global perspective.J Med Virol. 2022 Apr;94(4):1738-1744. doi: 10.1002/jmv.27524. Epub 2022 Jan 11. J Med Virol. 2022. PMID: 34905235
-
Omicron (B.1.1.529) - variant of concern - molecular profile and epidemiology: a mini review.Eur Rev Med Pharmacol Sci. 2021 Dec;25(24):8019-8022. doi: 10.26355/eurrev_202112_27653. Eur Rev Med Pharmacol Sci. 2021. PMID: 34982466 Review.
-
Molecular mimicry of host short linear motif-mediated interactions utilised by viruses for entry.Mol Biol Rep. 2023 May;50(5):4665-4673. doi: 10.1007/s11033-023-08389-2. Epub 2023 Apr 4. Mol Biol Rep. 2023. PMID: 37016039 Free PMC article. Review.
Cited by
-
Human TLS DNA polymerase: saviors or threats under replication stress?Mol Cell Biochem. 2025 May 23. doi: 10.1007/s11010-025-05291-2. Online ahead of print. Mol Cell Biochem. 2025. PMID: 40407973 Review.
-
Predicting Motif-Mediated Interactions Based on Viral Genomic Composition.Int J Mol Sci. 2025 Apr 13;26(8):3674. doi: 10.3390/ijms26083674. Int J Mol Sci. 2025. PMID: 40332242 Free PMC article.
-
Host-Pathogen Interaction 5.0.Int J Mol Sci. 2024 Dec 19;25(24):13596. doi: 10.3390/ijms252413596. Int J Mol Sci. 2024. PMID: 39769357 Free PMC article.
References
-
- Mészáros B., Sámano-Sánchez H., Alvarado-Valverde J., Čalyševa J., Martínez-Pérez E., Alves R., Shields D.C., Kumar M., Rippmann F., Chemes L.B., et al. Short Linear Motif Candidates in the Cell Entry System Used by SARS-CoV-2 and Their Potential Therapeutic Implications. 2021. [(accessed on 12 January 2021)]. Available online: http://elm.eu.org/ - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

