Annotation of Siberian Larch (Larix sibirica Ledeb.) Nuclear Genome-One of the Most Cold-Resistant Tree Species in the Only Deciduous GENUS in Pinaceae
- PMID: 35956540
- PMCID: PMC9370799
- DOI: 10.3390/plants11152062
Annotation of Siberian Larch (Larix sibirica Ledeb.) Nuclear Genome-One of the Most Cold-Resistant Tree Species in the Only Deciduous GENUS in Pinaceae
Abstract
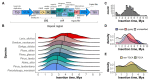
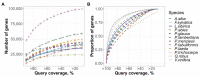
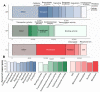
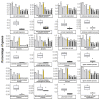
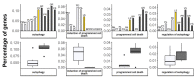
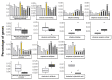
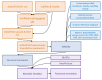
The recent release of the nuclear, chloroplast and mitochondrial genome assemblies of Siberian larch (Larix sibirica Ledeb.), one of the most cold-resistant tree species in the only deciduous genus of Pinaceae, with seasonal senescence and a rot-resistant valuable timber widely used in construction, greatly contributed to the development of genomic resources for the larch genus. Here, we present an extensive repeatome analysis and the first annotation of the draft nuclear Siberian larch genome assembly. About 66% of the larch genome consists of highly repetitive elements (REs), with the likely wave of retrotransposons insertions into the larch genome estimated to occur 4-5 MYA. In total, 39,370 gene models were predicted, with 87% of them having homology to the Arabidopsis-annotated proteins and 78% having at least one GO term assignment. The current state of the genome annotations allows for the exploration of the gymnosperm and angiosperm species for relative gene abundance in different functional categories. Comparative analysis of functional gene categories across different angiosperm and gymnosperm species finds that the Siberian larch genome has an overabundance of genes associated with programmed cell death (PCD), autophagy, stress hormone biosynthesis and regulatory pathways; genes that may play important roles in seasonal senescence and stress response to extreme cold in larch. Despite being incomplete, the draft assemblies and annotations of the conifer genomes are at a point of development where they now represent a valuable source for further genomic, genetic and population studies.
Keywords: RNA-seq; Siberian larch; angiosperms; annotation; conifer; deciduous; genome; gymnosperms; microsatellites; repeats; seasonal senescence; transcriptome; transposons.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- McLoughlin S. Gymnosperms. In: Alderton D., Elias S.A., editors. Encyclopedia of Geology. 2nd ed. Volume 3. Elsevier; Amsterdam, The Netherlands: 2021. pp. 476–500. - DOI
-
- Brenner E.D., Stevenson D. Using Genomics to Study Evolutionary Origins of Seeds. In: Williams C.G., editor. Landscapes, Genomics and Transgenic Conifers. Managing Forest Ecosystems. Volume 9. Springer; Dordrecht, The Netherlands: 2006. pp. 85–106. - DOI
-
- Soltis P.S., Soltis D.E., Savolainen V., Crane P.R., Barraclough T.G. Rate heterogeneity among lineages of tracheophytes: Integration of molecular and fossil data and evidence for molecular living fossils. Proc. Natl. Acad. Sci. USA. 2002;99:4430–4435. doi: 10.1073/pnas.032087199. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

