Conformational Dynamics of Human ALKBH2 Dioxygenase in the Course of DNA Repair as Revealed by Stopped-Flow Fluorescence Spectroscopy
- PMID: 35956910
- PMCID: PMC9370705
- DOI: 10.3390/molecules27154960
Conformational Dynamics of Human ALKBH2 Dioxygenase in the Course of DNA Repair as Revealed by Stopped-Flow Fluorescence Spectroscopy
Abstract
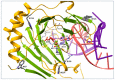
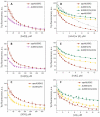
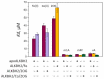
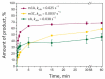
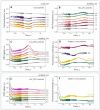
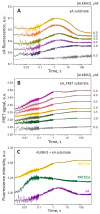
Elucidation of physicochemical mechanisms of enzymatic processes is one of the main tasks of modern biology. High efficiency and selectivity of enzymatic catalysis are mostly ensured by conformational dynamics of enzymes and substrates. Here, we applied a stopped-flow kinetic analysis based on fluorescent spectroscopy to investigate mechanisms of conformational transformations during the removal of alkylated bases from DNA by ALKBH2, a human homolog of Escherichia coli AlkB dioxygenase. This enzyme protects genomic DNA against various alkyl lesions through a sophisticated catalytic mechanism supported by a cofactor (Fe(II)), a cosubstrate (2-oxoglutarate), and O2. We present here a comparative study of conformational dynamics in complexes of the ALKBH2 protein with double-stranded DNA substrates containing N1-methyladenine, N3-methylcytosine, or 1,N6-ethenoadenine. By means of fluorescent labels of different types, simultaneous detection of conformational transitions in the protein globule and DNA substrate molecule was performed. Fitting of the kinetic curves by a nonlinear-regression method yielded a molecular mechanism and rate constants of its individual steps. The results shed light on overall conformational dynamics of ALKBH2 and damaged DNA during the catalytic cycle.
Keywords: DNA methylation; DNA repair; FRET analysis; aminopurine; conformational dynamics; dioxygenase ALKBH2; fluorescent spectroscopy; pre-steady-state kinetics; stopped-flow.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
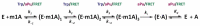
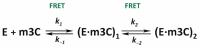

References
-
- Ringvoll J., Nordstrand L.M., Vagbo C.B., Talstad V., Reite K., Aas P.A., Lauritzen K.H., Liabakk N.B., Bjork A., Doughty R.W., et al. Repair deficient mice reveal mABH2 as the primary oxidative demethylase for repairing 1meA and 3meC lesions in DNA. Embo J. 2006;25:2189–2198. doi: 10.1038/sj.emboj.7601109. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

