Steroidogenic Factor 1 Regulates Transcription of the Inhibin B Coreceptor in Pituitary Gonadotrope Cells
- PMID: 35957608
- PMCID: PMC9761571
- DOI: 10.1210/endocr/bqac131
Steroidogenic Factor 1 Regulates Transcription of the Inhibin B Coreceptor in Pituitary Gonadotrope Cells
Abstract
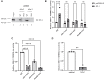
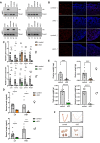
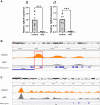
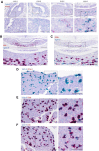
The inhibins control reproduction by suppressing follicle-stimulating hormone synthesis in pituitary gonadotrope cells. The newly discovered inhibin B coreceptor, TGFBR3L, is selectively and highly expressed in gonadotropes in both mice and humans. Here, we describe our initial characterization of mechanisms controlling cell-specific Tgfbr3l/TGFBR3L transcription. We identified two steroidogenic factor 1 (SF-1 or NR5A1) cis-elements in the proximal Tgfbr3l promoter in mice. SF-1 induction of murine Tgfbr3l promoter-reporter activity was inhibited by mutations in one or both sites in heterologous cells. In homologous cells, mutation of these cis-elements or depletion of endogenous SF-1 similarly decreased reporter activity. We observed nearly identical results when using a human TGFBR3L promoter-reporter. The Tgfbr3l gene was tightly compacted and Tgfbr3l mRNA expression was essentially absent in gonadotropes of SF-1 (Nr5a1) conditional knockout mice. During murine embryonic development, Tgfbr3l precedes Nr5a1 expression, though the two transcripts are fully colocalized by embryonic day 18.5 and thereafter. Collectively, these data indicate that SF-1 directly regulates Tgfbr3l/TGFBR3L transcription and is required for postnatal expression of the gene in gonadotropes.
Keywords: cell line; inhibin; knockout mouse; pituitary; receptor; transcription.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Endocrine Society. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures


Similar articles
-
Gonadotropin-releasing hormone regulates transcription of the inhibin B co-receptor, TGFBR3L, via early growth response one.J Biol Chem. 2025 Apr;301(4):108405. doi: 10.1016/j.jbc.2025.108405. Epub 2025 Mar 14. J Biol Chem. 2025. PMID: 40090584 Free PMC article.
-
TGFBR3L is an inhibin B co-receptor that regulates female fertility.Sci Adv. 2021 Dec 17;7(51):eabl4391. doi: 10.1126/sciadv.abl4391. Epub 2021 Dec 15. Sci Adv. 2021. PMID: 34910520 Free PMC article.
-
Betaglycan (TGFBR3) Functions as an Inhibin A, but Not Inhibin B, Coreceptor in Pituitary Gonadotrope Cells in Mice.Endocrinology. 2018 Dec 1;159(12):4077-4091. doi: 10.1210/en.2018-00770. Endocrinology. 2018. PMID: 30364975 Free PMC article.
-
A Tale of Two Proteins: Betaglycan, IGSF1, and the Continuing Search for the Inhibin B Receptor.Trends Endocrinol Metab. 2020 Jan;31(1):37-45. doi: 10.1016/j.tem.2019.08.014. Epub 2019 Oct 22. Trends Endocrinol Metab. 2020. PMID: 31648935 Review.
-
[An ambiguous role of steroidogenic factor 1 in the rat GnRH receptor gene expression. Lessons from transgenic mice].J Soc Biol. 2004;198(1):73-9. J Soc Biol. 2004. PMID: 15146959 Review. French.
Cited by
-
ZEB1 Inhibits LHβ Subunit Transcription When Overexpressed, but Is Dispensable for LH Synthesis in Mice.Endocrinology. 2024 Aug 27;165(10):bqae116. doi: 10.1210/endocr/bqae116. Endocrinology. 2024. PMID: 39248143 Free PMC article.
-
Intronic Enhancer Is Essential for Nr5a1 Expression in the Pituitary Gonadotrope and for Postnatal Development of Male Reproductive Organs in a Mouse Model.Int J Mol Sci. 2022 Dec 22;24(1):192. doi: 10.3390/ijms24010192. Int J Mol Sci. 2022. PMID: 36613635 Free PMC article.
-
Gonadotropin-releasing hormone regulates transcription of the inhibin B co-receptor, TGFBR3L, via early growth response one.J Biol Chem. 2025 Apr;301(4):108405. doi: 10.1016/j.jbc.2025.108405. Epub 2025 Mar 14. J Biol Chem. 2025. PMID: 40090584 Free PMC article.
References
-
- Simoni M, Weinbauer GF, Gromoll J, Nieschlag E. Role of FSH in male gonadal function. Ann Endocrinol (Paris). 1999;60(2):102‐106. - PubMed
-
- Woodruff TK, D’Agostino J, Schwartz NB, Mayo KE. Dynamic changes in inhibin messenger RNAs in rat ovarian follicles during the reproductive cycle. Science. 1988;239(4845):1296‐1299. - PubMed
-
- Woodruff TK, Meunier H, Jones PB, Hsueh AJ, Mayo KE. Rat inhibin: molecular cloning of alpha- and beta-subunit complementary deoxyribonucleic acids and expression in the ovary. Mol Endocrinol. 1987;1(8):561‐568. - PubMed

