Identification of novel biomarkers in septic cardiomyopathy via integrated bioinformatics analysis and experimental validation
- PMID: 35957694
- PMCID: PMC9358039
- DOI: 10.3389/fgene.2022.929293
Identification of novel biomarkers in septic cardiomyopathy via integrated bioinformatics analysis and experimental validation
Abstract
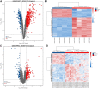
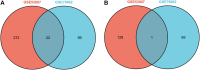
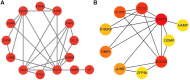
Purpose: Septic cardiomyopathy (SCM) is an important world public health problem with high morbidity and mortality. It is necessary to identify SCM biomarkers at the genetic level to identify new therapeutic targets and strategies. Method: DEGs in SCM were identified by comprehensive bioinformatics analysis of microarray datasets (GSE53007 and GSE79962) downloaded from the GEO database. Subsequently, bioinformatics analysis was used to conduct an in-depth exploration of DEGs, including GO and KEGG pathway enrichment analysis, PPI network construction, and key gene identification. The top ten Hub genes were identified, and then the SCM model was constructed by treating HL-1 cells and AC16 cells with LPS, and these top ten Hub genes were examined using qPCR. Result: STAT3, SOCS3, CCL2, IL1R2, JUNB, S100A9, OSMR, ZFP36, and HAMP were significantly elevated in the established SCM cells model. Conclusion: After bioinformatics analysis and experimental verification, it was demonstrated that STAT3, SOCS3, CCL2, IL1R2, JUNB, S100A9, OSMR, ZFP36, and HAMP might play important roles in SCM.
Keywords: biomarkers; differentially expressed genes; integrated bioinformatics analysis; sepsis; septic cardiomyopathy.
Copyright © 2022 Lu, Hu, Qiu, Zou and Xu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Chen L., Long X., Xu Q., Tan J., Wang G., Cao Y., et al. (2020a). Elevated serum levels of S100A8/A9 and HMGB1 at hospital admission are correlated with inferior clinical outcomes in COVID-19 patients.Research Support, Non-U.S. Gov't]. J. Artic. Mol. Immunol. 17 (9), 992–994. 10.1038/s41423-020-0492-x - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

