DNA damage repair-related gene signature predicts prognosis and indicates immune cell infiltration landscape in skin cutaneous melanoma
- PMID: 35957812
- PMCID: PMC9361349
- DOI: 10.3389/fendo.2022.882431
DNA damage repair-related gene signature predicts prognosis and indicates immune cell infiltration landscape in skin cutaneous melanoma
Abstract
Background: DNA damage repair plays an important role in the onset and progression of cancers and its resistance to treatment therapy. This study aims to assess the prognostic potential of DNA damage repair markers in skin cutaneous melanoma (SKCM).
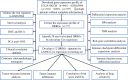
Method: In this study, we have analyzed the gene expression profiles being downloaded from TCGA, GTEx, and GEO databases. We sequentially used univariate and LASSO Cox regression analyses to screen DNA repair genes associated with prognosis. Then, we have conducted a multivariate regression analysis to construct the prognostic profile of DNA repair-related genes (DRRGs). The risk coefficient is used to calculate the risk scores and divide the patients into two cohorts. Additionally, we validated our prognosis model on an external cohort as well as evaluated the link between immune response and the DRRGs prognostic profiles. The risk signature is compared to immune cell infiltration, chemotherapy, and immune checkpoint inhibitors (ICIs) treatment.
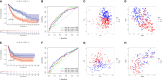
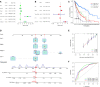
Results: An analysis using LASSO-Cox stepwise regression established a prognostic signature consisting of twelve DRRGs with strong predictive ability. Disease-specific survival (DSS) is found to be lower among high-risk patients group as compared to low-risk patients. The signature may be employed as an independent prognostic predictor after controlling for clinicopathological factors, as demonstrated by validation on one external GSE65904 cohort. A strong correlation is also found between the risk score and the immune microenvironment, along with the infiltrating immune cells, and ICIs key molecules. The gene enrichment analysis results indicate a wide range of biological activities and pathways to be exhibited by high-risk groups. Furthermore, Cisplatin exhibited a considerable response sensitivity in low-risk groups as opposed to the high-risk incidents, while docetaxel exhibited a considerable response sensitivity in high-risk groups.
Conclusions: Our findings provide a thorough investigation of DRRGs to develop an DSS-related prognostic indicator which may be useful in forecasting SKCM progression and enabling more enhanced clinical benefits from immunotherapy.
Keywords: DNA damage repair; immunotherapy; prognostic factor; skin cutaneous melanoma; tumor microenvironment.
Copyright © 2022 Liang, Mai, Mai, Chen and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Analysis on tumor immune microenvironment and construction of a prognosis model for immune-related skin cutaneous melanoma.Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2023 May 28;48(5):671-681. doi: 10.11817/j.issn.1672-7347.2023.230069. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2023. PMID: 37539569 Free PMC article. Chinese, English.
-
Development and validation of an immune gene set-based prognostic signature in cutaneous melanoma.Future Oncol. 2021 Nov;17(31):4115-4129. doi: 10.2217/fon-2021-0104. Epub 2021 Jul 22. Future Oncol. 2021. PMID: 34291650
-
The fatty acid-related gene signature stratifies poor prognosis patients and characterizes TIME in cutaneous melanoma.J Cancer Res Clin Oncol. 2024 Jan 27;150(2):40. doi: 10.1007/s00432-023-05580-7. J Cancer Res Clin Oncol. 2024. PMID: 38279987 Free PMC article.
-
Emerging prognostic biomarkers in advanced cutaneous melanoma: a literature update.Expert Rev Mol Diagn. 2024 Jan-Feb;24(1-2):49-66. doi: 10.1080/14737159.2024.2314574. Epub 2024 Feb 9. Expert Rev Mol Diagn. 2024. PMID: 38334382 Review.
-
DNA repair deficiency and the immune microenvironment: A pathways perspective.DNA Repair (Amst). 2024 Jan;133:103594. doi: 10.1016/j.dnarep.2023.103594. Epub 2023 Nov 13. DNA Repair (Amst). 2024. PMID: 37980867 Free PMC article. Review.
Cited by
-
The DNA damage repair-related lncRNAs signature predicts the prognosis and immunotherapy response in gastric cancer.Front Immunol. 2023 Jun 29;14:1117255. doi: 10.3389/fimmu.2023.1117255. eCollection 2023. Front Immunol. 2023. PMID: 37457685 Free PMC article.
-
Comprehensive investigation of matrix metalloproteinases in skin cutaneous melanoma: diagnostic, prognostic, and therapeutic insights.Sci Rep. 2025 Jan 16;15(1):2152. doi: 10.1038/s41598-025-85887-2. Sci Rep. 2025. PMID: 39820824 Free PMC article.
-
A novel DNA damage repair-related gene signature predicting survival, immune infiltration and drug sensitivity in cervical cancer based on single cell sequencing.Front Immunol. 2023 Jun 28;14:1198391. doi: 10.3389/fimmu.2023.1198391. eCollection 2023. Front Immunol. 2023. PMID: 37449209 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

