Bacterial Signal Peptides- Navigating the Journey of Proteins
- PMID: 35957980
- PMCID: PMC9360617
- DOI: 10.3389/fphys.2022.933153
Bacterial Signal Peptides- Navigating the Journey of Proteins
Abstract
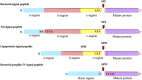
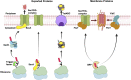
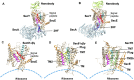
In 1971, Blobel proposed the first statement of the Signal Hypothesis which suggested that proteins have amino-terminal sequences that dictate their export and localization in the cell. A cytosolic binding factor was predicted, and later the protein conducting channel was discovered that was proposed in 1975 to align with the large ribosomal tunnel. The 1975 Signal Hypothesis also predicted that proteins targeted to different intracellular membranes would possess distinct signals and integral membrane proteins contained uncleaved signal sequences which initiate translocation of the polypeptide chain. This review summarizes the central role that the signal peptides play as address codes for proteins, their decisive role as targeting factors for delivery to the membrane and their function to activate the translocation machinery for export and membrane protein insertion. After shedding light on the navigation of proteins, the importance of removal of signal peptide and their degradation are addressed. Furthermore, the emerging work on signal peptidases as novel targets for antibiotic development is described.
Keywords: SecA; SecYEG translocase; Tat pathway; YidC; antibiotic targets; protein transport; signal peptidase; signal peptide.
Copyright © 2022 Kaushik, He and Dalbey.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


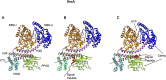




Similar articles
-
Signal Peptide Hydrophobicity Modulates Interaction with the Twin-Arginine Translocase.mBio. 2017 Aug 1;8(4):e00909-17. doi: 10.1128/mBio.00909-17. mBio. 2017. PMID: 28765221 Free PMC article.
-
Protein Transport Across the Bacterial Plasma Membrane by the Sec Pathway.Protein J. 2019 Jun;38(3):262-273. doi: 10.1007/s10930-019-09841-8. Protein J. 2019. PMID: 31134461 Review.
-
SecA alone can promote protein translocation and ion channel activity: SecYEG increases efficiency and signal peptide specificity.J Biol Chem. 2011 Dec 30;286(52):44702-9. doi: 10.1074/jbc.M111.300111. Epub 2011 Oct 27. J Biol Chem. 2011. PMID: 22033925 Free PMC article.
-
Promiscuous targeting of polytopic membrane proteins to SecYEG or YidC by the Escherichia coli signal recognition particle.Mol Biol Cell. 2012 Feb;23(3):464-79. doi: 10.1091/mbc.E11-07-0590. Epub 2011 Dec 7. Mol Biol Cell. 2012. PMID: 22160593 Free PMC article.
-
The Sec translocase.Biochim Biophys Acta. 2011 Mar;1808(3):851-65. doi: 10.1016/j.bbamem.2010.08.016. Epub 2010 Aug 27. Biochim Biophys Acta. 2011. PMID: 20801097 Review.
Cited by
-
Signal Peptides: From Molecular Mechanisms to Applications in Protein and Vaccine Engineering.Biomolecules. 2025 Jun 18;15(6):897. doi: 10.3390/biom15060897. Biomolecules. 2025. PMID: 40563537 Free PMC article. Review.
-
Key contributions of a glycolipid to membrane protein integration.Proc Jpn Acad Ser B Phys Biol Sci. 2024;100(7):387-413. doi: 10.2183/pjab.100.026. Proc Jpn Acad Ser B Phys Biol Sci. 2024. PMID: 39085064 Free PMC article. Review.
-
N-Terminal Sequences of Signal Peptides Assuming Critical Roles in Expression of Heterologous Proteins in Bacillus subtilis.Microorganisms. 2024 Jun 23;12(7):1275. doi: 10.3390/microorganisms12071275. Microorganisms. 2024. PMID: 39065044 Free PMC article.
-
Investigating in vivo Mycobacterium avium subsp. paratuberculosis microevolution and mixed strain infections.Microbiol Spectr. 2023 Aug 16;11(5):e0171623. doi: 10.1128/spectrum.01716-23. Online ahead of print. Microbiol Spectr. 2023. PMID: 37584606 Free PMC article.
-
Advancements in Escherichia coli secretion systems for enhanced recombinant protein production.World J Microbiol Biotechnol. 2025 Mar 3;41(3):90. doi: 10.1007/s11274-025-04302-0. World J Microbiol Biotechnol. 2025. PMID: 40025370 Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

