Molecular mechanism of hyperactive tooth root formation in oculo-facio-cardio-dental syndrome
- PMID: 35957990
- PMCID: PMC9359619
- DOI: 10.3389/fphys.2022.946282
Molecular mechanism of hyperactive tooth root formation in oculo-facio-cardio-dental syndrome
Abstract
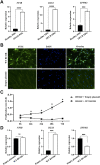
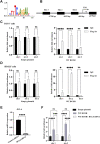
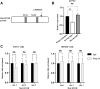
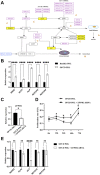
Mutations in the B-cell lymphoma 6 (BCL6) interacting corepressor (BCOR) cause oculo-facio-cardio-dental (OFCD) syndrome, a rare X-linked dominant condition that includes dental radiculomegaly among other characteristics. BCOR regulates downstream genes via BCL6 as a transcriptional corepressor. However, the molecular mechanism underlying the occurrence of radiculomegaly is still unknown. Thus, this study was aimed at identifying BCOR-regulated genetic pathways in radiculomegaly. The microarray profile of affected tissues revealed that the gene-specific transcriptional factors group, wherein nucleus factor 1B, distal-less homeobox 5, and zinc finger protein multitype 2 (ZFPM2) were the most upregulated, was significantly expressed in periodontal ligament (PDL) cells of the diseased patient with a frameshift mutation (c.3668delC) in BCOR. Wild-type BCOR overexpression in human periodontal ligament fibroblasts cells significantly hampered cellular proliferation and ZFPM2 mRNA downregulation. Promoter binding assays showed that wild-type BCOR was recruited in the BCL6 binding of the ZFPM2 promoter region after immunoprecipitation, while mutant BCOR, which was the same genotype as of our patient, failed to recruit these promoter regions. Knockdown of ZFPM2 expression in mutant PDL cells significantly reduced cellular proliferation as well as mRNA expression of alkaline phosphatase, an important marker of odontoblasts and cementoblasts. Collectively, our findings suggest that BCOR mutation-induced ZFPM2 regulation via BCL6 possibly contributes to hyperactive root formation in OFCD syndrome. Clinical data from patients with rare genetic diseases may aid in furthering the understanding of the mechanism controlling the final root length.
Keywords: BCL6; BCOR; OFCD syndrome; ZFPM2; radiculomegaly.
Copyright © 2022 Min Soe, Ogawa and Moriyama.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Nonsense-mediated mRNA decay affects hyperactive root formation in oculo-facio-cardio-dental syndrome via up-frameshift protein 1.J Oral Biosci. 2024 Mar;66(1):225-231. doi: 10.1016/j.job.2024.01.008. Epub 2024 Jan 19. J Oral Biosci. 2024. PMID: 38244688
-
Radiculomegaly as a key clinical feature in oculo-facio-cardio-dental (OFCD) syndrome: a case report with a novel truncating variant in BCOR gene.Cardiol Young. 2024 Dec;34(12):2606-2609. doi: 10.1017/S104795112402660X. Epub 2024 Oct 11. Cardiol Young. 2024. PMID: 39390895
-
Nuclear import of transcriptional corepressor BCOR occurs through interaction with karyopherin α expressed in human periodontal ligament.Biochem Biophys Res Commun. 2018 Dec 9;507(1-4):67-73. doi: 10.1016/j.bbrc.2018.10.158. Epub 2018 Nov 2. Biochem Biophys Res Commun. 2018. PMID: 30396568
-
Radiculomegaly: a case report of this rare dental finding with review of the associated oculo-facio-cardio-dental syndrome.Oral Surg Oral Med Oral Pathol Oral Radiol. 2018 Oct;126(4):e220-e227. doi: 10.1016/j.oooo.2018.02.011. Epub 2018 Feb 28. Oral Surg Oral Med Oral Pathol Oral Radiol. 2018. PMID: 29574060 Review.
-
Oculo-facio-cardio-dental (OFCD) syndrome.J Orofac Orthop. 1998;59(3):178-85. doi: 10.1007/BF01317179. J Orofac Orthop. 1998. PMID: 9640004 Review. English, German.
Cited by
-
[Oculo-facio-cardio-dental syndrome caused by BCOR gene mutations: a case report].Zhongguo Dang Dai Er Ke Za Zhi. 2023 Feb 15;25(2):202-204. doi: 10.7499/j.issn.1008-8830.2210045. Zhongguo Dang Dai Er Ke Za Zhi. 2023. PMID: 36854698 Free PMC article. Chinese.
References
-
- Fossett N., Tevosian S. G., Gajewski K., Zhang Q., Orkin S. H., Schulz R. A., et al. (2001). The Friend of GATA proteins U-shaped, FOG-1, and FOG-2 function as negative regulators of blood, heart, and eye development in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 98 (13), 7342–7347. 10.1073/pnas.131215798 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

