Phylogenomic analysis of the genus Leuconostoc
- PMID: 35958134
- PMCID: PMC9358442
- DOI: 10.3389/fmicb.2022.897656
Phylogenomic analysis of the genus Leuconostoc
Abstract
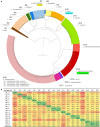
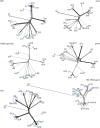
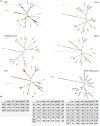
Leuconostoc is a genus of saccharolytic heterofermentative lactic acid bacteria that inhabit plant-derived matrices and a variety of fermented foods (dairy products, dough, milk, vegetables, and meats), contributing to desired fermentation processes or playing a role in food spoilage. At present, the genus encompasses 17 recognized species. In total, 216 deposited genome sequences of Leuconostoc were analyzed, to check the delineation of species and to infer their evolutive genealogy utilizing a minimum evolution tree of Average Nucleotide Identity (ANI) and the core genome alignment. Phylogenomic relationships were compared to those obtained from the analysis of 16S rRNA, pheS, and rpoA genes. All the phylograms were subjected to split decomposition analysis and their topologies were compared to check the ambiguities in the inferred phylogenesis. The minimum evolution ANI tree exhibited the most similar topology with the core genome tree, while single gene trees were less adherent and provided a weaker phylogenetic signal. In particular, the 16S rRNA gene failed to resolve several bifurcations and Leuconostoc species. Based on an ANI threshold of 95%, the organization of the genus Leuconostoc could be amended, redefining the boundaries of the species L. inhae, L. falkenbergense, L. gelidum, L. lactis, L. mesenteroides, and L. pseudomesenteroides. Two strains currently recognized as L. mesenteroides were split into a separate lineage representing a putative species (G16), phylogenetically related to both L. mesenteroides (G18) and L. suionicum (G17). Differences among the four subspecies of L. mesenteroides were not pinpointed by ANI or by the conserved genes. The strains of L. pseudomesenteroides were ascribed to two putative species, G13 and G14, the former including also all the strains presently belonging to L. falkenbergense. L. lactis was split into two phylogenetically related lineages, G9 and G10, putatively corresponding to separate species and both including subgroups that may correspond to subspecies. The species L. gelidum and L. gasicomitatum were closely related but separated into different species, the latter including also L. inhae strains. These results, integrating information of ANI, core genome, and housekeeping genes, complemented the taxonomic delineation with solid information on the phylogenetic lineages evolved within the genus Leuconostoc.
Keywords: 16S rRNA gene; Leuconostoc; average nucletide identity (ANI); biopreservatives; cosmeceutics; phylogenomics.
Copyright © 2022 Raimondi, Candeliere, Amaretti, Costa, Vertuani, Spampinato and Rossi.
Figures

Similar articles
-
Leuconostoc falkenbergense sp. nov., isolated from a lactic culture, fermentating string beans and traditional yogurt.Int J Syst Evol Microbiol. 2021 Jan;71(1). doi: 10.1099/ijsem.0.004602. Epub 2020 Dec 9. Int J Syst Evol Microbiol. 2021. PMID: 33295855
-
A proposal of Leuconostoc mesenteroides subsp. jonggajibkimchii subsp. nov. and reclassification of Leuconostoc mesenteroides subsp. suionicum (Gu et al., 2012) as Leuconostoc suionicum sp. nov. based on complete genome sequences.Int J Syst Evol Microbiol. 2017 Jul;67(7):2225-2230. doi: 10.1099/ijsem.0.001930. Epub 2017 Jul 3. Int J Syst Evol Microbiol. 2017. PMID: 28671527
-
Reclassification of Leuconostoc gasicomitatum as Leuconostoc gelidum subsp. gasicomitatum comb. nov., description of Leuconostoc gelidum subsp. aenigmaticum subsp. nov., designation of Leuconostoc gelidum subsp. gelidum subsp. nov. and emended description of Leuconostoc gelidum.Int J Syst Evol Microbiol. 2014 Apr;64(Pt 4):1290-1295. doi: 10.1099/ijs.0.058263-0. Epub 2014 Jan 15. Int J Syst Evol Microbiol. 2014. PMID: 24431060
-
Bacteriophages of leuconostoc, oenococcus, and weissella.Front Microbiol. 2014 Apr 28;5:186. doi: 10.3389/fmicb.2014.00186. eCollection 2014. Front Microbiol. 2014. PMID: 24817864 Free PMC article. Review.
-
Lactic acid bacteria of foods and their current taxonomy.Int J Food Microbiol. 1997 Apr 29;36(1):1-29. doi: 10.1016/s0168-1605(96)01233-0. Int J Food Microbiol. 1997. PMID: 9168311 Review.
Cited by
-
The Antibiotic Resistome and Its Association with Bacterial Communities in Raw Camel Milk from Altay Xinjiang.Foods. 2023 Oct 26;12(21):3928. doi: 10.3390/foods12213928. Foods. 2023. PMID: 37959048 Free PMC article.
-
Isolation and Characterization of Lactic Acid Bacteria With Probiotic Attributes From Different Parts of the Gastrointestinal Tract of Free-living Wild Boars in Hungary.Probiotics Antimicrob Proteins. 2024 Aug;16(4):1221-1239. doi: 10.1007/s12602-023-10113-2. Epub 2023 Jun 23. Probiotics Antimicrob Proteins. 2024. PMID: 37353593 Free PMC article.
-
Host genetics and gut microbiota synergistically regulate feed utilization in egg-type chickens.J Anim Sci Biotechnol. 2024 Sep 9;15(1):123. doi: 10.1186/s40104-024-01076-7. J Anim Sci Biotechnol. 2024. PMID: 39245742 Free PMC article.
-
Draft genome sequence of Leuconostoc falkenbergense isolated from naturally fermented buffalo milk curd.Microbiol Resour Announc. 2024 Apr 11;13(5):e0014824. doi: 10.1128/mra.00148-24. Online ahead of print. Microbiol Resour Announc. 2024. PMID: 38602401 Free PMC article.
-
The Metabolism of Leuconostoc Genus Decoded by Comparative Genomics.Microorganisms. 2024 Jul 20;12(7):1487. doi: 10.3390/microorganisms12071487. Microorganisms. 2024. PMID: 39065255 Free PMC article.
References
-
- Ahmadi-Ashtiani H. R., Baldisserotto A., Cesa E., Manfredini S., Zadeh H. S., Gorab M. G., et al. . (2020). Microbial biosurfactants as key multifunctional ingredients for sustainable cosmetics. Cosmetics 7, 46. 10.3390/COSMETICS7020046 - DOI
-
- Bello S., Rudra B., Gupta R. S. (2022). Phylogenomic and comparative genomic analyses of Leuconostocaceae species: identification of molecular signatures specific for the genera Leuconostoc, Fructobacillus and Oenococcus and proposal for a novel genus Periweissella gen. nov. Int. J. Syst. Evol. Microbiol. 72, 005284. 10.1099/ijsem.0.005284 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

