Integrative analysis of transcriptome and miRNAome reveals molecular mechanisms regulating pericarp thickness in sweet corn during kernel development
- PMID: 35958194
- PMCID: PMC9361504
- DOI: 10.3389/fpls.2022.945379
Integrative analysis of transcriptome and miRNAome reveals molecular mechanisms regulating pericarp thickness in sweet corn during kernel development
Abstract
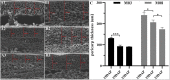
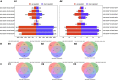
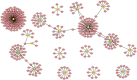
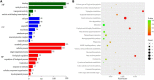
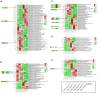
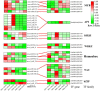
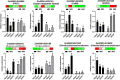
Pericarp thickness affects the edible quality of sweet corn (Zea mays L. saccharata Sturt.). Therefore, breeding varieties with a thin pericarp is important for the quality breeding of sweet corn. However, the molecular mechanisms underlying the pericarp development remain largely unclear. We performed an integrative analysis of mRNA and miRNA sequencing to elucidate the genetic mechanism regulating pericarp thickness during kernel development (at 15 days, 19 days, and 23 days after pollination) of two sweet corn inbred lines with different pericarp thicknesses (M03, with a thinner pericarp and M08, with a thicker pericarp). A total of 2,443 and 1,409 differentially expressed genes (DEGs) were identified in M03 and M08, respectively. Our results indicate that phytohormone-mediated programmed cell death (PCD) may play a critical role in determining pericarp thickness in sweet corn. Auxin (AUX), gibberellin (GA), and brassinosteroid (BR) signal transduction may indirectly mediate PCD to regulate pericarp thickness in M03 (the thin pericarp variety). In contrast, abscisic acid (ABA), cytokinin (CK), and ethylene (ETH) signaling may be the key regulators of pericarp PCD in M08 (the thick pericarp variety). Furthermore, 110 differentially expressed microRNAs (DEMIs) and 478 differentially expressed target genes were identified. miRNA164-, miRNA167-, and miRNA156-mediated miRNA-mRNA pairs may participate in regulating pericarp thickness. The expression results of DEGs were validated by quantitative real-time PCR. These findings provide insights into the molecular mechanisms regulating pericarp thickness and propose the objective of breeding sweet corn varieties with a thin pericarp.
Keywords: kernel development; miRNAome; pericarp thickness; sweet corn; transcriptome.
Copyright © 2022 Xiong, Pei, Zhang, Ren, Ma, Tang and Huang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
QTL mapping and transcriptome analysis identify candidate genes regulating pericarp thickness in sweet corn.BMC Plant Biol. 2020 Mar 14;20(1):117. doi: 10.1186/s12870-020-2295-8. BMC Plant Biol. 2020. PMID: 32171234 Free PMC article.
-
Root Transcriptome Analysis Identifies Salt-Tolerance Genes in Sweet Corn Chromosome Segment Substitution Lines (CSSLs).Plants (Basel). 2025 May 31;14(11):1687. doi: 10.3390/plants14111687. Plants (Basel). 2025. PMID: 40508362 Free PMC article.
-
Genome-Wide Transcriptome Analysis Revealing the Genes Related to Sugar Metabolism in Kernels of Sweet Corn.Metabolites. 2022 Dec 12;12(12):1254. doi: 10.3390/metabo12121254. Metabolites. 2022. PMID: 36557292 Free PMC article.
-
Physiological and Molecular Characteristics of Southern Leaf Blight Resistance in Sweet Corn Inbred Lines.Int J Mol Sci. 2022 Sep 6;23(18):10236. doi: 10.3390/ijms231810236. Int J Mol Sci. 2022. PMID: 36142144 Free PMC article.
-
Regulation of Phytohormones on the Growth and Development of Plant Root Hair.Front Plant Sci. 2022 Mar 24;13:865302. doi: 10.3389/fpls.2022.865302. eCollection 2022. Front Plant Sci. 2022. PMID: 35401627 Free PMC article. Review.
Cited by
-
A transcriptomic evaluation of the mechanism of programmed cell death of the replaceable bud in Chinese chestnut.Open Life Sci. 2023 Jul 7;18(1):20220635. doi: 10.1515/biol-2022-0635. eCollection 2023. Open Life Sci. 2023. PMID: 37426617 Free PMC article.
-
Comparative Transcriptome Analysis Reveals Key Functions of MiMYB Gene Family in Macadamia Nut Pericarp Formation.Int J Mol Sci. 2024 Jun 21;25(13):6840. doi: 10.3390/ijms25136840. Int J Mol Sci. 2024. PMID: 38999950 Free PMC article.
-
Genetic analysis and QTL mapping for pericarp thickness in maize (Zea mays L.).BMC Plant Biol. 2024 Apr 25;24(1):338. doi: 10.1186/s12870-024-05052-1. BMC Plant Biol. 2024. PMID: 38664642 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

