Mutations in OsDET1, OsCOP10, and OsDDB1 confer embryonic lethality and alter flavonoid accumulation in Rice (Oryza sativa L.) seed
- PMID: 35958215
- PMCID: PMC9358687
- DOI: 10.3389/fpls.2022.952856
Mutations in OsDET1, OsCOP10, and OsDDB1 confer embryonic lethality and alter flavonoid accumulation in Rice (Oryza sativa L.) seed
Abstract
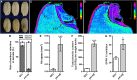
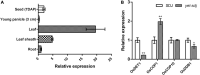
Morphological and biochemical changes accompanying embryogenesis and seed development are crucial for plant survival and crop productivity. Here, we identified a novel yellowish-pericarp embryo lethal (yel) mutant of the japonica rice cultivar Sindongjin (Oryza sativa L.), namely, yel-sdj. Seeds of the yel-sdj mutant showed a yellowish pericarp and black embryo, and were embryonic lethal. Compared with wild-type seeds, the yel-sdj mutant seeds exhibited significantly reduced grain size, grain weight, and embryo weight, and a remarkably lower rate of embryo retention in kernels subjected to milling. However, the volume of air space between embryo and endosperm, density of embryo, and total phenolic content (TPC) and antioxidant activity of mature grains were significantly higher in the yel-sdj mutant than in the wild type. Genetic analysis and mapping revealed that the yel-sdj mutant was non-allelic to the oscop1 null mutants yel-hc, yel-cc, and yel-sk, and its phenotype was controlled by a single recessive gene, LOC_Os01g01484, an ortholog of Arabidopsis thaliana DE-ETIOLATED 1 (DET1). The yel-sdj mutant carried a 7 bp deletion in the second exon of OsDET1. Seeds of the osdet1 knockout mutant, generated via CRISPR/Cas9-based gene editing, displayed the yel mutant phenotype. Consistent with the fact that OsDET1 interacts with CONSTITUTIVE PHOTOMORPHOGENIC 10 (OsCOP10) and UV-DAMAGED DNA BINDING PROTEIN 1 (OsDDB1) to form the COP10-DET1-DDB1 (CDD), seeds of oscop10 and osddb1 knockout mutants also showed the yel phenotype. These findings will enhance our understanding of the functional roles of OsDET1 and the CDD complex in embryogenesis and flavonoid biosynthesis in rice seeds.
Keywords: CDD complex; CRISPR/Cas9; OsDET1; embryo development; rice (Oryza sativa); yellowish-pericarp embryo lethal (yel) mutant.
Copyright © 2022 Kim, Lee, Nam, Lee, Seo, Lee, Cho, Kwon and Koh.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
OsCOP1 regulates embryo development and flavonoid biosynthesis in rice (Oryza sativa L.).Theor Appl Genet. 2021 Aug;134(8):2587-2601. doi: 10.1007/s00122-021-03844-9. Epub 2021 May 5. Theor Appl Genet. 2021. PMID: 33950284 Free PMC article.
-
The De-Etiolated 1 Homolog of Arabidopsis Modulates the ABA Signaling Pathway and ABA Biosynthesis in Rice.Plant Physiol. 2016 Jun;171(2):1259-76. doi: 10.1104/pp.16.00059. Epub 2016 May 2. Plant Physiol. 2016. PMID: 27208292 Free PMC article.
-
Comparative analysis of metabolite profiling and free radical scavenging activity in phenotypic variants of OsCOP1 colored rice mutant seed.Food Chem. 2023 Nov 1;425:136465. doi: 10.1016/j.foodchem.2023.136465. Epub 2023 May 27. Food Chem. 2023. PMID: 37276671
-
Mutation of OsDET1 increases chlorophyll content in rice.Plant Sci. 2013 Sep;210:241-9. doi: 10.1016/j.plantsci.2013.06.003. Epub 2013 Jun 13. Plant Sci. 2013. PMID: 23849131
-
OsLFR is essential for early endosperm and embryo development by interacting with SWI/SNF complex members in Oryza sativa.Plant J. 2020 Nov;104(4):901-916. doi: 10.1111/tpj.14967. Epub 2020 Sep 7. Plant J. 2020. PMID: 32808364
Cited by
-
Genome-Wide Association Study Reveals the Genetic Basis of Total Flavonoid Content in Brown Rice.Genes (Basel). 2023 Aug 25;14(9):1684. doi: 10.3390/genes14091684. Genes (Basel). 2023. PMID: 37761824 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

