Weighted Gene Coexpression Network Analysis Identifies Crucial Genes Involved in Coronary Atherosclerotic Heart Disease
- PMID: 35958279
- PMCID: PMC9363224
- DOI: 10.1155/2022/6971238
Weighted Gene Coexpression Network Analysis Identifies Crucial Genes Involved in Coronary Atherosclerotic Heart Disease
Abstract
Background: Coronary atherosclerotic heart disease (CHD) is a lethal disease with an unstated pathogenic mechanism. Therefore, it is urgent to develop innovative strategies to ameliorate the outcome of CHD patients and explore novel biomarkers connected to the pathogenicity of CHD.
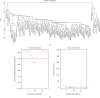
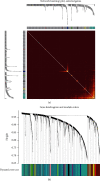
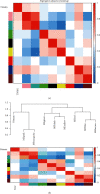
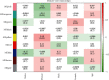
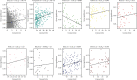
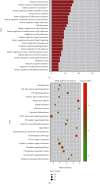
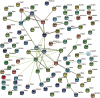
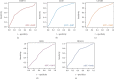
Methods: The weighted gene coexpression network analysis (WGCNA) was carried out on a coronary atherosclerosis dataset GSE90074 to determine the crucial modules and hub genes for their prospective relationship to CHD. After the different modules associated with CHD have been identified, the Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enriched pathway analyses were conducted. The protein-protein interaction (PPI) network was thereafter performed for the critical module using STRING and Cytoscape.
Results: The yellow module was recognized as the most critical module associated with CHD. The enriched pathways in the yellow module included those related to inflammatory response, positive regulation of extracellular signal-regulated kinase1/2 (ERK1/2) cascade, lipid catabolic process, cellular response to oxidative stress, apoptotic pathway, and NF-kappa B pathway. Further CytoHubba analysis revealed the top five hub genes (MMP14, CD28, CaMK4, RGS1, and DDAH1) associated with CHD development.
Conclusions: The current study provides the prognosis, novel hub genes, and signaling pathways for treating coronary atherosclerosis. However, their potential biological roles require deeper investigation.
Copyright © 2022 Jinli Bao.
Conflict of interest statement
The author declares no conflict of interest.
Figures
Similar articles
-
Weighted gene co-expression network analysis identifies specific modules and hub genes related to coronary artery disease.Sci Rep. 2021 Mar 23;11(1):6711. doi: 10.1038/s41598-021-86207-0. Sci Rep. 2021. PMID: 33758323 Free PMC article.
-
Identification of vital modules and genes associated with heart failure based on weighted gene coexpression network analysis.ESC Heart Fail. 2022 Apr;9(2):1370-1379. doi: 10.1002/ehf2.13827. Epub 2022 Feb 6. ESC Heart Fail. 2022. PMID: 35128826 Free PMC article.
-
Identifying Obstructive Sleep Apnea Syndrome-Associated Genes and Pathways through Weighted Gene Coexpression Network Analysis.Comput Math Methods Med. 2022 Jan 29;2022:3993509. doi: 10.1155/2022/3993509. eCollection 2022. Comput Math Methods Med. 2022. PMID: 35132330 Free PMC article.
-
Integration of Gene Expression Profile Data of Human Epicardial Adipose Tissue from Coronary Artery Disease to Verification of Hub Genes and Pathways.Biomed Res Int. 2019 Nov 27;2019:8567306. doi: 10.1155/2019/8567306. eCollection 2019. Biomed Res Int. 2019. PMID: 31886261 Free PMC article.
-
Identification of the pivotal role of SPP1 in kidney stone disease based on multiple bioinformatics analysis.BMC Med Genomics. 2022 Jan 11;15(1):7. doi: 10.1186/s12920-022-01157-4. BMC Med Genomics. 2022. PMID: 35016690 Free PMC article.
Cited by
-
The Impact of the Neutrophil-to-Lymphocyte Ratio on In-Hospital Outcomes in Patients With Acute ST-Segment Elevation Myocardial Infarction.Cureus. 2024 Feb 18;16(2):e54418. doi: 10.7759/cureus.54418. eCollection 2024 Feb. Cureus. 2024. PMID: 38375058 Free PMC article.
-
G protein regulation by RGS proteins in the pathophysiology of dilated cardiomyopathy.Am J Physiol Heart Circ Physiol. 2025 Feb 1;328(2):H348-H360. doi: 10.1152/ajpheart.00653.2024. Epub 2025 Jan 7. Am J Physiol Heart Circ Physiol. 2025. PMID: 39772618 Free PMC article. Review.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

