BARX2/FOXA1/HK2 axis promotes lung adenocarcinoma progression and energy metabolism reprogramming
- PMID: 35958341
- PMCID: PMC9359959
- DOI: 10.21037/tlcr-22-465
BARX2/FOXA1/HK2 axis promotes lung adenocarcinoma progression and energy metabolism reprogramming
Abstract
Background: Metabolic reprogramming is an emerging cancer feature that has recently drawn special attention since it promotes tumor cell growth and proliferation. However, the mechanism of the Warburg effect is still largely unknown. This research aimed to reveal the effects of BarH-like homeobox 2 (BARX2) in regulating tumor progression and glucose metabolism in lung adenocarcinoma (LUAD).
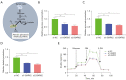
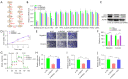
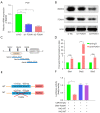
Methods: Expression of BARX2 was measured by quantitative real-time polymerase chain reaction (qRT-PCR) in LUAD cell line and tissues, and the tumor-promoting function of BARX2 in LUAD cells was detected in vitro and in vivo xenograft models. The metabolic effects of BARX2 were examined by detecting glucose uptake, the production levels of lactate and pyruvate, and the extracellular acidification rate (ECAR). Chromatin immunoprecipitation (ChIP) assay and luciferase reporter gene assay were used to identify the underlying molecular mechanism of BARX2 regulation of HK2. Further studies showed that transcription factor FOXA1 directly interacts with BARX2 and promotes the transcriptional activity of BARX2.
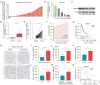
Results: BARX2 was remarkably up-regulated in LUAD tissues and positively linked to advanced clinical stage and poor prognosis. In vitro and in vivo data indicated ectopic expression of BARX2 enhanced cell proliferation and tumorigenesis, whereas BARX2 knockdown suppressed these effects. Metabolic-related experiments showed BARX2 promoted the reprogramming of glucose metabolism. Mechanistically, the BARX2/FOXA1/HK2 axis promoted LUAD progression and energy metabolism reprogramming.
Conclusions: In summary, our research first defined BARX2 as a tumor-promoting factor in LUAD and that it may act as a novel prognostic biomarker and new therapeutic target for the disease.
Keywords: BARX2; HK2; Transcription factor; glucose metabolism; lung adenocarcinoma (LUAD).
2022 Translational Lung Cancer Research. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-465/coif). Prof. RAdM received research grant from CNPQ Brazil, Libss, Pfizer; royalties from Springer; consulting fee from Takeda; speaker fee from Merck, Pfizer, Novartis, Eurofarma, MSD, Bayer, Astrazenenca; supporting for attending meetings and stock from Merck. Prof. RAdM also serves in Advisory board for European School of Oncology and Brazilian Society of Cancerology. ACR received Royalties from educational material for thymic tumors; payment from Honorarium for grand rounds at NY Langone; and Travel support for visiting professorship at NYU Langone; none of them are pertinent to this publication. The other authors have no conflicts of interest to declare.
Figures



Similar articles
-
Transcription Factor FOXA1 Facilitates Glycolysis and Proliferation of Lung Adenocarcinoma via Activation of TEX19.Mol Biotechnol. 2024 Aug;66(8):2144-2154. doi: 10.1007/s12033-023-00848-2. Epub 2023 Aug 22. Mol Biotechnol. 2024. PMID: 37606876
-
Downregulation of BarH-like homeobox 2 promotes cell proliferation, migration and aerobic glycolysis through Wnt/β-catenin signaling, and predicts a poor prognosis in non-small cell lung carcinoma.Thorac Cancer. 2018 Mar;9(3):390-399. doi: 10.1111/1759-7714.12593. Epub 2018 Jan 17. Thorac Cancer. 2018. PMID: 29341468 Free PMC article.
-
circFBXW7 attenuates malignant progression in lung adenocarcinoma by sponging miR-942-5p.Transl Lung Cancer Res. 2021 Mar;10(3):1457-1473. doi: 10.21037/tlcr-21-230. Transl Lung Cancer Res. 2021. PMID: 33889522 Free PMC article.
-
TRIB3 Promotes Lung Adenocarcinoma Progression via an Enhanced Warburg Effect.Cancer Manag Res. 2020 Dec 23;12:13195-13206. doi: 10.2147/CMAR.S287956. eCollection 2020. Cancer Manag Res. 2020. PMID: 33380827 Free PMC article.
-
Correlation of Glucose Metabolism with Cancer and Intervention with Traditional Chinese Medicine.Evid Based Complement Alternat Med. 2022 Oct 14;2022:2192654. doi: 10.1155/2022/2192654. eCollection 2022. Evid Based Complement Alternat Med. 2022. PMID: 36276846 Free PMC article. Review.
Cited by
-
Restoring BARX2 in OSCC reverses partial EMT and suppresses metastasis through miR-186-5p/miR-378a-3p-dependent SERPINE2 inhibition.Oncogene. 2024 Jun;43(25):1941-1954. doi: 10.1038/s41388-024-03053-w. Epub 2024 May 8. Oncogene. 2024. PMID: 38719950
-
Unlocking the secrets of glucose metabolism reprogramming: the role in pulmonary diseases.Front Pharmacol. 2025 Aug 13;16:1551452. doi: 10.3389/fphar.2025.1551452. eCollection 2025. Front Pharmacol. 2025. PMID: 40880642 Free PMC article. Review.
-
Deciphering the role of PLCD3 in lung cancer: A gateway to glycolytic reprogramming via PKC-Rap1 activation.Heliyon. 2024 Aug 30;10(17):e37063. doi: 10.1016/j.heliyon.2024.e37063. eCollection 2024 Sep 15. Heliyon. 2024. PMID: 39296221 Free PMC article.
-
Pathogenic role of super-enhancers as potential therapeutic targets in lung cancer.Front Pharmacol. 2024 Apr 12;15:1383580. doi: 10.3389/fphar.2024.1383580. eCollection 2024. Front Pharmacol. 2024. PMID: 38681203 Free PMC article. Review.
-
Toward precision oncology in LUAD: a prognostic model using single-cell sequencing and WGCNA based on a disulfidptosis relative gene signature.Front Immunol. 2025 May 21;16:1581915. doi: 10.3389/fimmu.2025.1581915. eCollection 2025. Front Immunol. 2025. PMID: 40469299 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
