A signature based on m6A pattern and tumor microenvironment infiltration in clear cell renal cell carcinoma
- PMID: 35958449
- PMCID: PMC9360897
A signature based on m6A pattern and tumor microenvironment infiltration in clear cell renal cell carcinoma
Abstract
Background: RNA N6-methyladenosine (m6A) has been found to have a critical impact on clear cell renal cell carcinoma (ccRCC) by affecting the tumor microenvironment (TME) and immune cell (IC) infiltration and is related to the treatment and survival rate of patients with ccRCC. However, the mechanism of m6A in TME and IC infiltration remained unclear.
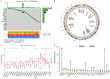
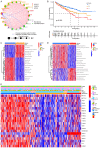
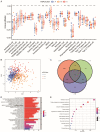
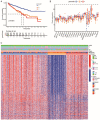
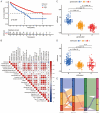
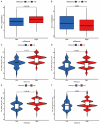
Methods: Nonnegative Matrix Factorization (NMF) clustering was performed on 650 ccRCC cases from the Cancer Genome Atlas (TCGA) and the Gene-Expression Omnibus (GEO) datasets. The immune infiltration was generated by the single-sample gene-set enrichment analysis (ssGSEA) algorithm. Survival analyses were performed using the Kaplan-Meier method, and the significance of the differences was determined using the log-rank test. The m6A score was constructed based on the expression of m6A regulators to quantify m6A modification. The package "survminer R" was employed to layer patients' low and high scores groups and predict the immunotherapy response.
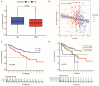
Results: Three different patterns of m6A modification were established, and significant differences in TME and IC infiltration features were found in these three patterns. Survival analysis demonstrated that m6A cluster A and m6A gene cluster A experienced a longer survival time. Evaluation of m6A modification patterns in individual tumors was initiated by the m6A score. The low m6A score subtype was characterized by increased tumor mutation burden (TMB) and immune infiltration, whereas a high m6A score with a lack of immune cell infiltration showed significantly better overall survival. m6A score was also associated with the expression of programmed cell death protein 1 (PD-L1) and cytotoxic T lymphocyte antigen 4 (CTLA-4). Patients in the high m6A score group had high PD-L1 expression and low CTLA-4 expression. Significant differences in prognosis were identified among types of different TMB and m6A scores, where low TMB and high m6A score had longer survival time.
Conclusions: This research indicated that m6A modification greatly affected TME and IC infiltration. Physicians can develop practical immunotherapy strategies for patients with ccRCC by evaluating m6A-associated genes.
Keywords: ccRCC; immunotherapy; m6A; tumor microenvironment; tumor mutation burden.
AJTR Copyright © 2022.
Conflict of interest statement
None.
Figures
References
-
- Williamson SR. Clear cell papillary renal cell carcinoma: an update after 15 years. Pathology. 2021;53:109–119. - PubMed
-
- Wettersten HI, Aboud OA, Lara PN Jr, Weiss RH. Metabolic reprogramming in clear cell renal cell carcinoma. Nat Rev Nephrol. 2017;13:410–419. - PubMed
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
