Identification of immune hub genes participating in the pathogenesis and progression of Vogt-Koyanagi-Harada disease
- PMID: 35958546
- PMCID: PMC9358976
- DOI: 10.3389/fimmu.2022.936707
Identification of immune hub genes participating in the pathogenesis and progression of Vogt-Koyanagi-Harada disease
Abstract
Background: Vogt-Koyanagi-Harada (VKH) disease is an autoimmune inflammatory disorder characterized by bilateral granulomatous uveitis. The objective of this study was to identify immune hub genes involved in the pathogenesis and progression of VKH disease.
Methods: High throughput sequencing data were downloaded from the Gene Expression Omnibus (GEO) and an immune dataset was downloaded from ImmPort. Immune differentially expressed genes (DEGs) were obtained from their intersection in the GEO and ImmPort datasets. Immune hub genes for VKH disease were selected through differential expression analyses, including Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), Disease Ontology (DO), protein-protein interaction (PPI) network, and clustering analyses. Confidence in the immune hub genes was subsequently validated using box plots and receiver operating characteristic (ROC) curves.
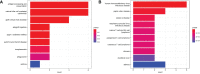
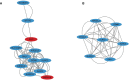
Results: A total of 254 DEGs were screened and after the intersection with ImmPort, 20 genes were obtained as immune DEGs. Functional enrichment analysis indicated that the key genes were mainly involved in several types of immune pathways (such as the lymphocyte mediated and leukocyte mediated immune responses, natural killer cell mediated cytotoxicity, and antigen binding) and immunodeficiency diseases. Following PPI network analysis, the top seven genes in cluster 1 were selected as potential immune hub genes in VKH. After evaluating the accuracy of the hub genes, one gene (GNLY) was excluded because its expression level was statistically similar in VKH patients and healthy controls. Finally, six immune hub genes, namely KLRC2, KLRC3 SH2D1B, GZMB, KIR2DL3, and KIR3DL2 were identified as playing important roles in the occurrence and development of VKH disease.
Conclusion: Six immune hub genes (KLRC2, KLRC3 SH2D1B, GZMB, KIR2DL3, and KIR3DL2) identified by our bioinformatics analyses may provide new diagnostic and therapeutic targets for VKH disease.
Keywords: Vogt-Koyanagi-Harada disease; functional enrichment analyses; immune hub genes; protein-protein interaction network analysis; receiver operating characteristic curves.
Copyright © 2022 Wang, Ju, Wang, Sun, Tang, Gao, Gu and Ji.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

