HTLV-1 intragenic viral enhancer influences immortalization phenotype in vitro, but is dispensable for persistence and disease development in animal models
- PMID: 35958554
- PMCID: PMC9359075
- DOI: 10.3389/fimmu.2022.954077
HTLV-1 intragenic viral enhancer influences immortalization phenotype in vitro, but is dispensable for persistence and disease development in animal models
Abstract
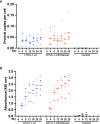
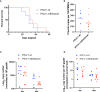
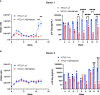
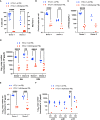
Human T-cell leukemia virus type 1 (HTLV-1) is the causative infectious agent of adult T-cell leukemia/lymphoma (ATL) and chronic neurological disease. The disparity between silenced sense transcription versus constitutively active antisense (Hbz) transcription from the integrated provirus is not fully understood. The presence of an internal viral enhancer has recently been discovered in the Tax gene near the 3' long terminal repeat (LTR) of HTLV-1. In vitro, this enhancer has been shown to bind SRF and ELK-1 host transcription factors, maintain chromatin openness and viral gene transcription, and induce aberrant host gene transcription near viral integration sites. However, the function of the viral enhancer in the context of early HTLV-1 infection events remains unknown. In this study, we generated a mutant Enhancer virus (mEnhancer) and evaluated its effects on HTLV-1-mediated in vitro immortalization, establishment of persistent infection with an in vivo rabbit model, and disease development in a humanized immune system (HIS) mouse model. The mEnhancer virus was able to establish persistent infection in rabbits, and there were no significant differences in proviral load or HTLV-1-specific antibody responses over a 25-week study. However, rabbits infected with the mEnhancer virus had significantly decreased sense and antisense viral gene expression at 12-weeks post-infection. HIS mice infected with wt or mEnhancer virus showed similar disease progression, proviral load, and viral gene expression. While mEnhancer virus was able to sufficiently immortalize primary T-lymphocytes in cell culture, the immortalized cells had an altered phenotype (CD8+ T-cells), decreased proviral load, decreased sense and anti-sense gene expression, and altered cell cycle progression compared to HTLV-1.wt immortalized cells (CD4+ T-cells). These results suggest that the HTLV-1 enhancer element alone does not determine persistence or disease development but plays a pivotal role in regulating viral gene expression.
Keywords: HTLV-1; enhancer; immortalization; persistence; retrovirus; transcription.
Copyright © 2022 Maksimova, Smith, Seth, Phelps, Niewiesk, Satou, Green and Panfil.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

