Establishment of a circular RNA regulatory stemness-related gene pair signature for predicting prognosis and therapeutic response in colorectal cancer
- PMID: 35958575
- PMCID: PMC9357884
- DOI: 10.3389/fimmu.2022.934124
Establishment of a circular RNA regulatory stemness-related gene pair signature for predicting prognosis and therapeutic response in colorectal cancer
Abstract
Background: Colorectal cancer (CRC) is a common malignant tumor of the digestive tract with a poor prognosis. Cancer stem cells (CSCs) affect disease outcomes and treatment responses in CRC. We developed a circular RNA (circRNA) regulatory stemness-related gene pair (CRSRGP) signature to predict CRC patient prognosis and treatment effects.
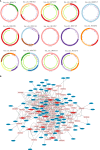
Methods: The circRNA, miRNA, and mRNA expression profiles and clinical information of CRC patients were obtained from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases. CRSRGPs were established based on stemness-related genes in the competing endogenous RNA (ceRNA) network. A CRSRGP signature was generated using the least absolute shrinkage and selection operator (Lasso) and Cox regression analysis of TCGA training set. The prognosis was predicted by generating a nomogram integrating the CRSRGP signature and clinicopathologic features. The model was validated in an external validation set (GSE17536). The antitumor drug sensitivity and immunotherapy responses of CRC patients in the high-risk group (HRG) and low-risk group (LRG) were evaluated by the pRRophetic algorithm and immune checkpoint analysis.
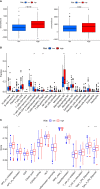
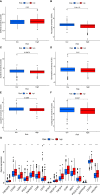
Results: We established an 18-CRSRGP signature to predict the prognosis and treatment responses of CRC patients. In the training and external validation sets, risk scores were used to categorize CRC patients into the HRG and LRG. The Kaplan-Meier analysis showed a poor prognosis for patients in the HRG and that subgroups with different clinical characteristics had significantly different prognoses. A multivariate Cox analysis revealed that the CRSRGP signature was an independent prognostic factor. The nomogram integrating clinical features and the CRSRGP signature efficiently predicted CRC patient prognosis, outperformed the current TNM staging system, and had improved practical clinical value. Anticancer drug sensitivity predictions revealed that the tumors of patients in the HRG were more sensitive to pazopanib, sunitinib, gemcitabine, lapatinib, and cyclopamine. Analysis of immune checkpoint markers demonstrated that patients in the HRG were more likely to benefit from immunotherapy.
Conclusion: An efficient, reliable tool for evaluating CRC patient prognosis and treatment response was established based on the 18-CRSRGP signature and nomogram.
Keywords: circRNA; colorectal cancer; immune; nomogram; stemness-related gene pair signature.
Copyright © 2022 Chen, Tang, Huang and Qiu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

