Prediction and Screening Model for Products Based on Fusion Regression and XGBoost Classification
- PMID: 35958779
- PMCID: PMC9357736
- DOI: 10.1155/2022/4987639
Prediction and Screening Model for Products Based on Fusion Regression and XGBoost Classification
Abstract
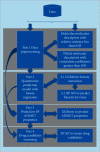
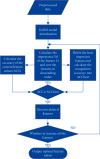
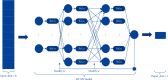
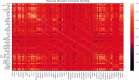
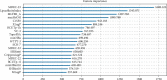
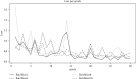
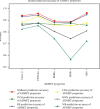
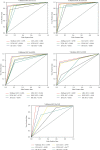
Performance prediction based on candidates and screening based on predicted performance value are the core of product development. For example, the performance prediction and screening of equipment components and parts are an important guarantee for the reliability of equipment products. The prediction and screening of drug bioactivity value and performance are the keys to pharmaceutical product development. The main reasons for the failure of pharmaceutical discovery are the low bioactivity of the candidate compounds and the deficiencies in their efficacy and safety, which are related to the absorption, distribution, metabolism, excretion, and toxicity (ADMET) of the compounds. Therefore, it is very necessary to quickly and effectively perform systematic bioactivity value prediction and ADMET property evaluation for candidate compounds in the early stage of drug discovery. In this paper, a data-driven pharmaceutical products screening prediction model is proposed to screen drug candidates with higher bioactivity value and better ADMET properties. First, a quantitative prediction method for bioactivity value is proposed using the fusion regression of LGBM and neural network based on backpropagation (BP-NN). Then, the ADMET properties prediction method is proposed using XGBoost. According to the predicted bioactivity value and ADMET properties, the BVAP method is defined to screen the drug candidates. And the screening model is validated on the dataset of antagonized Erα active compounds, in which the mean square error (MSE) of fusion regression is 1.1496, the XGBoost prediction accuracy of ADMET properties are 94.0% for Caco-2, 95.7% for CYP3A4, 89.4% for HERG, 88.6% for hob, and 96.2% for Mn. Compared with the commonly used methods for ADMET properties such as SVM, RF, KNN, LDA, and NB, the XGBoost in this paper has the highest prediction accuracy and AUC value, which has better guiding significance and can help screen pharmaceutical product candidates with good bioactivity, pharmacokinetic properties, and safety.
Copyright © 2022 Jiaju Wu et al.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures
Similar articles
-
Prediction of ADMET Properties of Anti-Breast Cancer Compounds Using Three Machine Learning Algorithms.Molecules. 2023 Mar 2;28(5):2326. doi: 10.3390/molecules28052326. Molecules. 2023. PMID: 36903569 Free PMC article.
-
A machine learning-based approach to ERα bioactivity and drug ADMET prediction.Front Genet. 2023 Jan 4;13:1087273. doi: 10.3389/fgene.2022.1087273. eCollection 2022. Front Genet. 2023. PMID: 36685926 Free PMC article.
-
Conformalized Graph Learning for Molecular ADMET Property Prediction and Reliable Uncertainty Quantification.J Chem Inf Model. 2024 Dec 9;64(23):8705-8717. doi: 10.1021/acs.jcim.4c01139. Epub 2024 Nov 21. J Chem Inf Model. 2024. PMID: 39571080
-
De-risking drug discovery with ADDME -- avoiding drug development mistakes early.Altern Lab Anim. 2009 Sep;37 Suppl 1:47-55. doi: 10.1177/026119290903701S10. Altern Lab Anim. 2009. PMID: 19807206 Review.
-
Improving ADMET Prediction Accuracy for Candidate Drugs: Factors to Consider in QSPR Modeling Approaches.Curr Top Med Chem. 2024;24(3):222-242. doi: 10.2174/0115680266280005231207105900. Curr Top Med Chem. 2024. PMID: 38083894 Review.
Cited by
-
Enhancing ERα-targeted compound efficacy in breast cancer threapy with ExplainableAI and GeneticAlgorithm.PLoS One. 2025 May 20;20(5):e0319673. doi: 10.1371/journal.pone.0319673. eCollection 2025. PLoS One. 2025. PMID: 40392928 Free PMC article.
-
Double-head transformer neural network for molecular property prediction.J Cheminform. 2023 Feb 23;15(1):27. doi: 10.1186/s13321-023-00700-4. J Cheminform. 2023. PMID: 36823530 Free PMC article.
-
Normalization and selecting non-differentially expressed genes improve machine learning modelling of cross-platform transcriptomic data.ArXiv [Preprint]. 2025 Jan 24:arXiv:2501.14248v1. ArXiv. 2025. Update in: Trans Artif Intell. 2025;1(1):5. doi: 10.53941/tai.2025.100005. PMID: 39975431 Free PMC article. Updated. Preprint.
-
Normalization and Selecting Non-Differentially Expressed Genes Improve Machine Learning Modelling of Cross-Platform Transcriptomic Data.Trans Artif Intell. 2025;1(1):5. doi: 10.53941/tai.2025.100005. Epub 2025 May 25. Trans Artif Intell. 2025. PMID: 40630982 Free PMC article.
-
Prediction of ADMET Properties of Anti-Breast Cancer Compounds Using Three Machine Learning Algorithms.Molecules. 2023 Mar 2;28(5):2326. doi: 10.3390/molecules28052326. Molecules. 2023. PMID: 36903569 Free PMC article.
References
-
- PhRMA. Biopharmaceutical Research Industry Profile . Washington, DC, USA: 2018. http://www.phrma.org/industrvprofile/2018/
-
- Gift S., Bader A. A low-cost, high-quality new drug discovery process using patient-derived induced pluripotent stem cells. Drug Discovery Today . 2015;20(1):37–49. - PubMed
-
- Haiping. Methods: 2019. IVS2vec: a tool of Inverse Virtual Screening based on word2vec and deep learning techniques. - PubMed
-
- Yang H. Study on the prediction and optimization method of ADMET properties of compounds . China: East China University of Science and Technology;
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

