Exploration of Potential Biomarkers and Immune Landscape for Hepatoblastoma: Evidence from Machine Learning Algorithm
- PMID: 35958911
- PMCID: PMC9357682
- DOI: 10.1155/2022/2417134
Exploration of Potential Biomarkers and Immune Landscape for Hepatoblastoma: Evidence from Machine Learning Algorithm
Retraction in
-
Retracted: Exploration of Potential Biomarkers and Immune Landscape for Hepatoblastoma: Evidence from Machine Learning Algorithm.Evid Based Complement Alternat Med. 2023 Oct 18;2023:9893765. doi: 10.1155/2023/9893765. eCollection 2023. Evid Based Complement Alternat Med. 2023. PMID: 37886423 Free PMC article.
Abstract
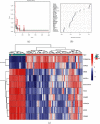
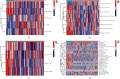
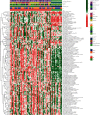
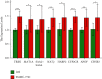
This study aimed to investigate the immune landscape in hepatoblastoma (HB) based on deconvolution methods and identify a biomarkers panel for diagnosis based on a machine learning algorithm. Firstly, we identified 277 differentially expressed genes (DEGs) and differentiated and functionally identified the modules in DEGs. The Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis and GO (gene ontology) were used to annotate these DEGs, and the results suggested that the occurrence of HB was related to DNA adducts, bile secretion, and metabolism of xenobiotics by cytochrome P450. We selected the top 10 genes for our final diagnostic panel based on the random forest tree method. Interestingly, TNFRSF19 and TOP2A were significantly down-regulated in normal samples, while other genes (TRIB1, MAT1A, SAA2-SAA4, NAT2, HABP2, CYP2CB, APOF, and CFHR3) were significantly down-regulated in HB samples. Finally, we constructed a neural network model based on the above hub genes for diagnosis. After cross-validation, the area under the ROC curve was close to 1 (AUC = 0.972), and the AUC of the validation set was 0.870. In addition, the results of single-sample gene-set enrichment analysis (ssGSEA) and deconvolution methods revealed a more active immune responses in the HB tissue. In conclusion, we have developed a robust biomarkers panel for HB patients.
Copyright © 2022 Peng Zhou et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures




Similar articles
-
Identification of Ten Core Hub Genes as Potential Biomarkers and Treatment Target for Hepatoblastoma.Front Oncol. 2021 Apr 1;11:591507. doi: 10.3389/fonc.2021.591507. eCollection 2021. Front Oncol. 2021. PMID: 33868991 Free PMC article.
-
Bioinformatics analysis of diagnostic biomarkers for Alzheimer's disease in peripheral blood based on sex differences and support vector machine algorithm.Hereditas. 2022 Oct 4;159(1):38. doi: 10.1186/s41065-022-00252-x. Hereditas. 2022. PMID: 36195955 Free PMC article.
-
Identification of immune- and autophagy-related genes and effective diagnostic biomarkers in endometriosis: a bioinformatics analysis.Ann Transl Med. 2022 Dec;10(24):1397. doi: 10.21037/atm-22-5979. Ann Transl Med. 2022. PMID: 36660690 Free PMC article.
-
Integrative Analysis of DNA Methylation and Gene Expression Profiling Data Reveals Candidate Methylation-Regulated Genes in Hepatoblastoma.Int J Gen Med. 2021 Dec 6;14:9419-9431. doi: 10.2147/IJGM.S331178. eCollection 2021. Int J Gen Med. 2021. PMID: 34908869 Free PMC article.
-
Establishment and Analysis of an Artificial Neural Network Model for Early Detection of Polycystic Ovary Syndrome Using Machine Learning Techniques.J Inflamm Res. 2023 Nov 29;16:5667-5676. doi: 10.2147/JIR.S438838. eCollection 2023. J Inflamm Res. 2023. PMID: 38050562 Free PMC article.
Cited by
-
Retracted: Exploration of Potential Biomarkers and Immune Landscape for Hepatoblastoma: Evidence from Machine Learning Algorithm.Evid Based Complement Alternat Med. 2023 Oct 18;2023:9893765. doi: 10.1155/2023/9893765. eCollection 2023. Evid Based Complement Alternat Med. 2023. PMID: 37886423 Free PMC article.
-
Phosphorylation-Mediated Activation of β-Catenin-TCF4-CEGRs/ALCDs Pathway Is an Essential Event in Development of Aggressive Hepatoblastoma.Cancers (Basel). 2022 Dec 9;14(24):6062. doi: 10.3390/cancers14246062. Cancers (Basel). 2022. PMID: 36551548 Free PMC article.
References
-
- Hager J., Sergi C. M. Hepatoblastoma. In: Sergi C. M., editor. Liver Cancer . Brisbane, Australia: Exon Publications; 2021.
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

