Long Noncoding RNA HLA Complex Group 18 Improves the Cell Proliferation of Myocardial Fibroblasts by Regulating the Hsa-microRNA-133a/Epidermal Growth Factor Receptor Axis
- PMID: 35958914
- PMCID: PMC9357715
- DOI: 10.1155/2022/2668239
Long Noncoding RNA HLA Complex Group 18 Improves the Cell Proliferation of Myocardial Fibroblasts by Regulating the Hsa-microRNA-133a/Epidermal Growth Factor Receptor Axis
Retraction in
-
Retracted: Long Noncoding RNA HLA Complex Group 18 Improves the Cell Proliferation of Myocardial Fibroblasts by Regulating the Hsa-microRNA-133a/Epidermal Growth Factor Receptor Axis.Evid Based Complement Alternat Med. 2023 Oct 4;2023:9818754. doi: 10.1155/2023/9818754. eCollection 2023. Evid Based Complement Alternat Med. 2023. PMID: 37829633 Free PMC article.
Abstract
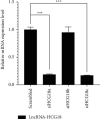
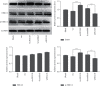
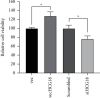
Hsa-microRNA (has-miR)-133a inactivates the extracellular signal-regulated kinase 1/2 (ERK1/2) pathway and suppresses the cell proliferation of myocardial fibroblasts by downregulation of the epidermal growth factor receptor (EGFR) expression. Bioinformatics analysis exhibits extended noncoding RNA HLA complex group 18 (lncRNA-HCG18) binds to hsa-miR-133a. The purpose of the current experiment is to explore whether lncRNA-HCG18 adsorbed hsa-miR-133a through sponging, resulting in decreased inhibition of hsa-miR-133a on EGFR and ultimately promoting the proliferation of myocardial fibroblasts. To verify and study the correlation and mechanism between lncRNA-HCG18, hsa-miR-133a, and their target genes. Firstly, after overexpression/silencing of lncRNA-HCG18 in myocardial fibroblasts, the level of hsa-miR-133a expression was evaluated by quantitative reverse transcription-polymerase chain reaction (RT-qPCR), and the EGFR, ERK1/2, and p-ERK1/2 expression levels were assessed by Western blotting to confirm that upregulation of EGFR and p-ERK1/2 protein levels by overexpression of lncRNA-HCG18, siRNA lncRNA-HCG18 (siHCG18) reduced the EGFR and p-ERK1/2 protein levels. Then, the luciferase reporter gene system was used to verify that lncRNA-HCG18 regulated EGFR expression by inhibiting the function of the hsa-miR-133a regulatory target gene. Then, a RAP assay was used to confirm that lncRNA-HCG18 interacted with hsa-miR-133a. Finally, the analysis of CCK-8 results indicated that the cell proliferation of myocardial fibroblasts was significantly reduced by siHCG18 while reversed by overexpression of lncRNA-HCG18. Thus, lncRNA-HCG18 inhibited cell viability of cardiac fibroblasts via the hsa-miR-133a/EGFR axis, which was regarded as a regulator of cell proliferation of cardiac fibroblasts in cardiovascular diseases.
Copyright © 2022 Huoshun Shi et al.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures




Similar articles
-
LncRNA HCG18 Promotes Osteosarcoma Cells Proliferation, Migration, and Invasion in by Regulating miR-34a/RUNX2 Pathway.Biochem Genet. 2023 Jun;61(3):1035-1049. doi: 10.1007/s10528-022-10294-5. Epub 2022 Nov 19. Biochem Genet. 2023. PMID: 36401685
-
lncRNA HLA Complex Group 18 (HCG18) Facilitated Cell Proliferation, Invasion, and Migration of Prostate Cancer Through Modulating miR-370-3p/DDX3X Axis.Reprod Sci. 2021 Dec;28(12):3406-3416. doi: 10.1007/s43032-021-00614-2. Epub 2021 Oct 27. Reprod Sci. 2021. PMID: 34708395
-
LncRNA HCG18 upregulates TRAF4/TRAF5 to facilitate proliferation, migration and EMT of epithelial ovarian cancer by targeting miR-29a/b.Mol Med. 2022 Jan 4;28(1):2. doi: 10.1186/s10020-021-00415-y. Mol Med. 2022. PMID: 34983361 Free PMC article.
-
Long non-coding RNA HCG18 promotes M1 macrophage polarization through regulating the miR-146a/TRAF6 axis, facilitating the progression of diabetic peripheral neuropathy.Mol Cell Biochem. 2021 Jan;476(1):471-482. doi: 10.1007/s11010-020-03923-3. Epub 2020 Sep 29. Mol Cell Biochem. 2021. PMID: 32996080
-
The role of lncRNA HCG18 in human diseases.Cell Biochem Funct. 2024 Mar;42(2):e3961. doi: 10.1002/cbf.3961. Cell Biochem Funct. 2024. PMID: 38425124 Review.
Cited by
-
Mechanism of action of non-coding RNAs and traditional Chinese medicine in myocardial fibrosis: Focus on the TGF-β/Smad signaling pathway.Front Pharmacol. 2023 Feb 9;14:1092148. doi: 10.3389/fphar.2023.1092148. eCollection 2023. Front Pharmacol. 2023. PMID: 36843918 Free PMC article. Review.
-
Retracted: Long Noncoding RNA HLA Complex Group 18 Improves the Cell Proliferation of Myocardial Fibroblasts by Regulating the Hsa-microRNA-133a/Epidermal Growth Factor Receptor Axis.Evid Based Complement Alternat Med. 2023 Oct 4;2023:9818754. doi: 10.1155/2023/9818754. eCollection 2023. Evid Based Complement Alternat Med. 2023. PMID: 37829633 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
