Two-step clustering-based pipeline for big dynamic functional network connectivity data
- PMID: 35958983
- PMCID: PMC9358255
- DOI: 10.3389/fnins.2022.895637
Two-step clustering-based pipeline for big dynamic functional network connectivity data
Abstract
Background: Dynamic functional network connectivity (dFNC) estimated from resting-state functional magnetic imaging (rs-fMRI) studies the temporally varying functional integration between brain networks. In a conventional dFNC pipeline, a clustering stage to summarize the connectivity patterns that are transiently but reliably realized over the course of a scanning session. However, identifying the right number of clusters (or states) through a conventional clustering criterion computed by running the algorithm repeatedly over a large range of cluster numbers is time-consuming and requires substantial computational power even for typical dFNC datasets, and the computational demands become prohibitive as datasets become larger and scans longer. Here we developed a new dFNC pipeline based on a two-step clustering approach to analyze large dFNC data without having access to huge computational power.
Methods: In the proposed dFNC pipeline, we implement two-step clustering. In the first step, we randomly use a sub-sample dFNC data and identify several sets of states at different model orders. In the second step, we aggregate all dFNC states estimated from all iterations in the first step and use this to identify the optimum number of clusters using the elbow criteria. Additionally, we use this new reduced dataset and estimate a final set of states by performing a second kmeans clustering on the aggregated dFNC states from the first k-means clustering. To validate the reproducibility of results in the new pipeline, we analyzed four dFNC datasets from the human connectome project (HCP).
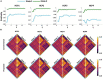
Results: We found that both conventional and proposed dFNC pipelines generate similar brain dFNC states across all four sessions with more than 99% similarity. We found that the conventional dFNC pipeline evaluates the clustering order and finds the final dFNC state in 275 min, while this process takes only 11 min for the proposed dFNC pipeline. In other words, the new pipeline is 25 times faster than the traditional method in finding the optimum number of clusters and finding the final dFNC states. We also found that the new method results in better clustering quality than the conventional approach (p < 0.001). We show that the results are replicated across four different datasets from HCP.
Conclusion: We developed a new analytic pipeline that facilitates the analysis of large dFNC datasets without having access to a huge computational power source. We validated the reproducibility of the result across multiple datasets.
Keywords: big data; dynamic functional network connectivity; human connectome project; kmeans clustering; reproducibility.
Copyright © 2022 Sendi, Salat, Miller and Calhoun.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- Béjar Alonso J. (2013). K-means vs Mini Batch K-Means: A Comparison. Available online at: http://hdl.handle.net/2117/23414 (accessed May, 2013).
LinkOut - more resources
Full Text Sources
Miscellaneous

