Identification of gut metabolites associated with Parkinson's disease using bioinformatic analyses
- PMID: 35959296
- PMCID: PMC9360421
- DOI: 10.3389/fnagi.2022.927625
Identification of gut metabolites associated with Parkinson's disease using bioinformatic analyses
Abstract
Background: Parkinson's disease (PD) is a common neurodegenerative disease affecting the movement of elderly patients. Environmental exposures are the risk factors for PD; however, gut environmental risk factors for PD are critically understudied. The proof-of-concept study is to identify gut metabolites in feces, as environmental exposure risk factors, that are associated with PD and potentially increase the risk for PD by using leverage of known toxicology results.
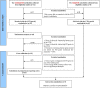
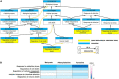
Materials and methods: We collected the data regarding the gut metabolites whose levels were significantly changed in the feces of patients with PD from the original clinical studies after searching the following databases: EBM Reviews, PubMed, Embase, MEDLINE, and Elsevier ClinicalKey. We further searched each candidate metabolite-interacting PD gene set by using the public Comparative Toxicogenomics Database (CTD), identified and validated gut metabolites associated with PD, and determined gut metabolites affecting specific biological functions and cellular pathways involved in PD by using PANTHER tools.
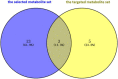
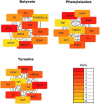
Results: Sixteen metabolites were identified and divided into the following main categories according to their structures and biological functions: alcohols (ethanol), amino acids (leucine, phenylalanine, pyroglutamic acid, glutamate, and tyrosine), short-chain fatty acids (propionate and butyrate), unsaturated fatty acids (linoleic acid and oleic acid), energy metabolism (lactate, pyruvate, and fumarate), vitamins (nicotinic acid and pantothenic acid), and choline metabolism (choline). Finally, a total of three identified metabolites, including butyrate, tyrosine, and phenylalanine, were validated that were associated with PD.
Conclusion: Our findings identified the gut metabolites that were highly enriched for PD genes and potentially increase the risk of developing PD. The identification of gut metabolite exposures can provide biomarkers for disease identification, facilitate an understanding of the relationship between gut metabolite exposures and response, and present an opportunity for PD prevention and therapies.
Keywords: Comparative Toxicogenomics Database; Parkinson’s disease; environmental exposures; gut metabolites; visual analyses.
Copyright © 2022 Yan, Feng, Zhou, Zhao, Xiao, Li and Shen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Gut Microbiota and Metabolome Alterations Associated with Parkinson's Disease.mSystems. 2020 Sep 15;5(5):e00561-20. doi: 10.1128/mSystems.00561-20. mSystems. 2020. PMID: 32934117 Free PMC article.
-
Alteration of Gut Microbial Metabolites in the Systemic Circulation of Patients with Parkinson's Disease.J Parkinsons Dis. 2022;12(4):1219-1230. doi: 10.3233/JPD-223179. J Parkinsons Dis. 2022. PMID: 35342048
-
Quantitative metabolomics of saliva using proton NMR spectroscopy in patients with Parkinson's disease and healthy controls.Neurol Sci. 2020 May;41(5):1201-1210. doi: 10.1007/s10072-019-04143-4. Epub 2020 Jan 3. Neurol Sci. 2020. PMID: 31897951
-
Potential roles of gut microbiota and microbial metabolites in Parkinson's disease.Ageing Res Rev. 2021 Aug;69:101347. doi: 10.1016/j.arr.2021.101347. Epub 2021 Apr 24. Ageing Res Rev. 2021. PMID: 33905953 Review.
-
Biomarker Research in Parkinson's Disease Using Metabolite Profiling.Metabolites. 2017 Aug 11;7(3):42. doi: 10.3390/metabo7030042. Metabolites. 2017. PMID: 28800113 Free PMC article. Review.
Cited by
-
Localized Pantothenic Acid (Vitamin B5) Reductions Present Throughout the Dementia with Lewy Bodies Brain.J Parkinsons Dis. 2024;14(5):965-976. doi: 10.3233/JPD-240075. J Parkinsons Dis. 2024. PMID: 38820022 Free PMC article.
-
Tyrosine metabolic reprogramming coordinated with the tricarboxylic acid cycle to drive glioma immune evasion by regulating PD-L1 expression.Ibrain. 2023 May 22;9(2):133-147. doi: 10.1002/ibra.12107. eCollection 2023 Summer. Ibrain. 2023. PMID: 37786553 Free PMC article.
-
From the Gut to the Brain: Is Microbiota a New Paradigm in Parkinson's Disease Treatment?Cells. 2024 Apr 30;13(9):770. doi: 10.3390/cells13090770. Cells. 2024. PMID: 38727306 Free PMC article. Review.
-
Causal association between phenylalanine and Parkinson's disease: a two-sample bidirectional mendelian randomization study.Front Genet. 2024 Jul 1;15:1322551. doi: 10.3389/fgene.2024.1322551. eCollection 2024. Front Genet. 2024. PMID: 39011398 Free PMC article.
-
Microbiota-Gut-Brain Axis in Age-Related Neurodegenerative Diseases.Curr Neuropharmacol. 2025;23(5):524-546. doi: 10.2174/1570159X23666241101093436. Curr Neuropharmacol. 2025. PMID: 39501955 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources

