Enterobacterial LPS-inducible LINC00152 is regulated by histone lactylation and promotes cancer cells invasion and migration
- PMID: 35959377
- PMCID: PMC9359126
- DOI: 10.3389/fcimb.2022.913815
Enterobacterial LPS-inducible LINC00152 is regulated by histone lactylation and promotes cancer cells invasion and migration
Abstract
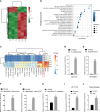
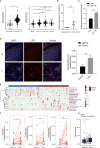
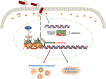
Gut microbes participate in pathogenesis by interacting with the host genome through epigenetic mechanisms, such as long non-coding RNAs. However, the mechanisms by which the microbiota induce expression alteration of long non-coding RNAs remains unclear. Here, we quantified the transcriptome alteration of human colon cell lines after being infected by a common enteric pathogen Salmonella typhimurium SL1344. We observed a widespread lncRNAs expression alteration. Among them, the elevated expression of LINC00152 was verified and proved to be induced by enteric bacteria-derived lipopolysaccharide (LPS). The inducible LINC00152 were found to inhibit Salmonella invasion and inflammation response. LINC00152 was overexpressed in tumors of the clinical CRC samples compared with adjacent normal tissues. Accordingly, we also demonstrated that overexpression of LINC00152 promoted the migration and invasion of colorectal cancer cells. Consistently, we observed an increased abundance of gram-negative bacteria and LPS in tumors tissue. Taken together, the above data implicated that enriched gram-negative bacteria in tumor tissue might promote tumor growth through modulating the expression of LINC00152. Furthermore, we demonstrated that LPS upregulated the expression of LINC00152 by introducing histone lactylation on its promoter and decreasing the binding efficiency of the repressor, YY1, to it. Our results provide new insights into how enterobacteria affect host epigenetics in human disease.
Keywords: colorectal cancer; enteric bacteria; histone lactylation; lipopolysaccharide; lncRNA.
Copyright © 2022 Wang, Liu, Xu, Wang, Wang, Zhang, Ni, Zhen, Xu, Liu, Fang, Huang and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Long intergenic non-coding RNA 00152 promotes lung adenocarcinoma proliferation via interacting with EZH2 and repressing IL24 expression.Mol Cancer. 2017 Jan 21;16(1):17. doi: 10.1186/s12943-017-0581-3. Mol Cancer. 2017. PMID: 28109288 Free PMC article.
-
Long Intergenic Noncoding RNA 00152 Promotes Glioma Cell Proliferation and Invasion by Interacting with MiR-16.Cell Physiol Biochem. 2018;46(3):1055-1064. doi: 10.1159/000488836. Epub 2018 Apr 13. Cell Physiol Biochem. 2018. PMID: 29669323
-
YY1-regulated LINC00152 promotes triple negative breast cancer progression by affecting on stability of PTEN protein.Biochem Biophys Res Commun. 2019 Feb 5;509(2):448-454. doi: 10.1016/j.bbrc.2018.12.074. Epub 2018 Dec 26. Biochem Biophys Res Commun. 2019. PMID: 30594392
-
Improved characterization of the relationship between long intergenic non-coding RNA Linc00152 and the occurrence and development of malignancies.Cancer Med. 2019 Aug;8(10):4722-4731. doi: 10.1002/cam4.2245. Epub 2019 Jul 4. Cancer Med. 2019. PMID: 31270960 Free PMC article. Review.
-
LINC00152: A pivotal oncogenic long non-coding RNA in human cancers.Cell Prolif. 2017 Aug;50(4):e12349. doi: 10.1111/cpr.12349. Epub 2017 May 2. Cell Prolif. 2017. PMID: 28464433 Free PMC article. Review.
Cited by
-
Lactylation in cancer: Mechanisms in tumour biology and therapeutic potentials.Clin Transl Med. 2024 Nov;14(11):e70070. doi: 10.1002/ctm2.70070. Clin Transl Med. 2024. PMID: 39456119 Free PMC article. Review.
-
Effects of Long Non-Coding RNAs Induced by the Gut Microbiome on Regulating the Development of Colorectal Cancer.Cancers (Basel). 2022 Nov 25;14(23):5813. doi: 10.3390/cancers14235813. Cancers (Basel). 2022. PMID: 36497293 Free PMC article. Review.
-
Histone lactylation: from tumor lactate metabolism to epigenetic regulation.Int J Biol Sci. 2024 Mar 3;20(5):1833-1854. doi: 10.7150/ijbs.91492. eCollection 2024. Int J Biol Sci. 2024. PMID: 38481814 Free PMC article. Review.
-
Lactate-induced protein lactylation in cancer: functions, biomarkers and immunotherapy strategies.Front Immunol. 2025 Jan 10;15:1513047. doi: 10.3389/fimmu.2024.1513047. eCollection 2024. Front Immunol. 2025. PMID: 39867891 Free PMC article. Review.
-
The role of lactate metabolism and lactylation in pulmonary arterial hypertension.Respir Res. 2025 Mar 12;26(1):99. doi: 10.1186/s12931-025-03163-3. Respir Res. 2025. PMID: 40075458 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

