Pan-cancer Landscape of the RUNX Protein Family Reveals their Potential as Carcinogenic Biomarkers and the Mechanisms Underlying their Action
- PMID: 35959452
- PMCID: PMC9328034
- DOI: 10.2478/jtim-2022-0013
Pan-cancer Landscape of the RUNX Protein Family Reveals their Potential as Carcinogenic Biomarkers and the Mechanisms Underlying their Action
Abstract
Background: The RUNX family of transcription factors plays an important regulatory role in tumor development. Although the importance of RUNX in certain cancer types is well known, the pan-cancer landscape remains unclear.
Materials and methods: Data from The Cancer Genome Atlas (TCGA) provides a pan-cancer overview of the RUNX genes. Hence, herein, we performed a pan-cancer analysis of abnormal RUNX expression and deciphered the potential regulatory mechanism. Specifically, we used TCGA multi-omics data combined with multiple online tools to analyze transcripts, genetic alterations, DNA methylation, clinical prognoses, miRNA networks, and potential target genes.
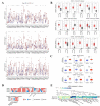
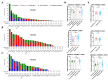
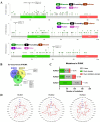
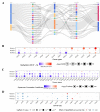
Results: RUNX genes are consistently overexpressed in esophageal, gastric, pancreatic, and pan-renal cancers. The total protein expression of RUNX1 in lung adenocarcinoma, kidney renal clear cell carcinoma (KIRC), and uterine corpus endometrial carcinoma (UCEC) is consistent with the mRNA expression results. Moreover, increased phosphorylation on the T14 and T18 residues of RUNX1 may represent potential pathogenic factors. The RUNX genes are significantly associated with survival in pan-renal cancer, brain lower-grade glioma, and uveal melanoma. Meanwhile, various mutations and posttranscriptional changes, including the RUNX1 D96 mutation in invasive breast carcinoma, the co-occurrence of RUNX gene mutations in UCEC, and methylation changes in the RUNX2 promoter in KIRC, may be associated with cancer development. Finally, analysis of epigenetic regulator co-expression, miRNA networks, and target genes revealed the carcinogenicity, abnormal expression, and direct regulation of RUNX genes.
Conclusions: We successfully analyzed the pan-cancer abnormal expression and prognostic value of RUNX genes, thereby providing potential biomarkers for various cancers. Further, mutations revealed via genetic alteration analysis may serve as a basis for personalized patient therapies.
Keywords: RUNX family; The Cancer Genome Atlas; pan-cancer analysis; prognosis; regulatory mechanism.
© 2022 Shen Pan, Siyu Sun, Bitian Liu, Yang Hou, published by Sciendo.
Figures





Similar articles
-
Identification of SHCBP1 as a potential biomarker involving diagnosis, prognosis, and tumor immune microenvironment across multiple cancers.Comput Struct Biotechnol J. 2022 Jun 18;20:3106-3119. doi: 10.1016/j.csbj.2022.06.039. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35782736 Free PMC article.
-
Pan-Cancer Analysis of PARP1 Alterations as Biomarkers in the Prediction of Immunotherapeutic Effects and the Association of Its Expression Levels and Immunotherapy Signatures.Front Immunol. 2021 Aug 31;12:721030. doi: 10.3389/fimmu.2021.721030. eCollection 2021. Front Immunol. 2021. PMID: 34531868 Free PMC article.
-
The panoramic picture of pepsinogen gene family with pan-cancer.Cancer Med. 2020 Dec;9(23):9064-9080. doi: 10.1002/cam4.3489. Epub 2020 Oct 17. Cancer Med. 2020. PMID: 33067881 Free PMC article.
-
The cancer driver genes IDH1/2, JARID1C/ KDM5C, and UTX/ KDM6A: crosstalk between histone demethylation and hypoxic reprogramming in cancer metabolism.Exp Mol Med. 2019 Jun 20;51(6):1-17. doi: 10.1038/s12276-019-0230-6. Exp Mol Med. 2019. PMID: 31221981 Free PMC article. Review.
-
CROX (Cluster Regulation of RUNX) as a Potential Novel Therapeutic Approach.Mol Cells. 2020 Feb 29;43(2):198-202. doi: 10.14348/molcells.2019.0268. Mol Cells. 2020. PMID: 31991534 Free PMC article. Review.
Cited by
-
TOM40 as a prognostic oncogene for oral squamous cell carcinoma prognosis.BMC Cancer. 2025 Jan 15;25(1):92. doi: 10.1186/s12885-024-13417-w. BMC Cancer. 2025. PMID: 39815211 Free PMC article.
-
Novel biomarkers: the RUNX family as prognostic predictors in colorectal cancer.Front Immunol. 2024 Dec 9;15:1430136. doi: 10.3389/fimmu.2024.1430136. eCollection 2024. Front Immunol. 2024. PMID: 39822248 Free PMC article.
-
Organoids to Remodel SARS-CoV-2 Research: Updates, Limitations and Perspectives.Aging Dis. 2023 Oct 1;14(5):1677-1699. doi: 10.14336/AD.2023.0209. Aging Dis. 2023. PMID: 37196111 Free PMC article. Review.
-
A pan-cancer analysis of anti-proliferative protein family genes for therapeutic targets in cancer.Sci Rep. 2023 Dec 7;13(1):21607. doi: 10.1038/s41598-023-48961-1. Sci Rep. 2023. PMID: 38062199 Free PMC article.
-
Unveiling clinical significance and tumor immune landscape of CXCL12 in bladder cancer: Insights from multiple omics analysis.Chin J Cancer Res. 2023 Dec 30;35(6):686-701. doi: 10.21147/j.issn.1000-9604.2023.06.12. Chin J Cancer Res. 2023. PMID: 38204439 Free PMC article.
References
-
- Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M. The Human Transcription Factors. Cell. 2018;172:650–65. et al. - PubMed
-
- Papavassiliou KA, Papavassiliou AG. Transcription Factor Drug Targets. J Cell Biochem. 2016;117:2693–6. - PubMed
-
- Ito Y, Bae SC, Chuang LS. The RUNX family: developmental regulators in cancer. Nat Rev Cancer. 2015;15:81–95. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
