Chanalyzer: A Computational Geometry Approach for the Analysis of Protein Channel Shape and Dynamics
- PMID: 35959458
- PMCID: PMC9358003
- DOI: 10.3389/fmolb.2022.933924
Chanalyzer: A Computational Geometry Approach for the Analysis of Protein Channel Shape and Dynamics
Abstract
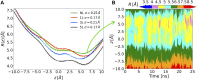
Morphological analysis of protein channels is a key step for a thorough understanding of their biological function and mechanism. In this respect, molecular dynamics (MD) is a very powerful tool, enabling the description of relevant biological events at the atomic level, which might elude experimental observations, and pointing to the molecular determinants thereof. In this work, we present a computational geometry-based approach for the characterization of the shape and dynamics of biological ion channels or pores to be used in combination with MD trajectories. This technique relies on the earliest works of Edelsbrunner and on the NanoShaper software, which makes use of the alpha shape theory to build the solvent-excluded surface of a molecular system in an aqueous solution. In this framework, a channel can be simply defined as a cavity with two entrances on the opposite sides of a molecule. Morphological characterization, which includes identification of the main axis, the corresponding local radius, and the detailed description of the global shape of the cavity, is integrated with a physico-chemical description of the surface facing the pore lumen. Remarkably, the possible existence or temporary appearance of fenestrations from the channel interior towards the outer lipid matrix is also accounted for. As a test case, we applied the present approach to the analysis of an engineered protein channel, the mechanosensitive channel of large conductance.
Keywords: alpha shapes theory; channel and pore characterization; computational geometry; ion channels; molecular dynamics; molecular surface; skeletonization.
Copyright © 2022 Raffo, Gagliardi, Fugacci, Sagresti, Grandinetti, Brancato, Biasotti and Rocchia.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Darden T., York D., Pedersen L. (1993). Particle Mesh Ewald: AnN⋅Log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 98, 10089–10092. 10.1063/1.464397 - DOI
LinkOut - more resources
Full Text Sources
Research Materials

