Distinct composition and amplification dynamics of transposable elements in sacred lotus (Nelumbo nucifera Gaertn.)
- PMID: 35959634
- PMCID: PMC9804982
- DOI: 10.1111/tpj.15938
Distinct composition and amplification dynamics of transposable elements in sacred lotus (Nelumbo nucifera Gaertn.)
Abstract
Sacred lotus (Nelumbo nucifera Gaertn.) is a basal eudicot plant with a unique lifestyle, physiological features, and evolutionary characteristics. Here we report the unique profile of transposable elements (TEs) in the genome, using a manually curated repeat library. TEs account for 59% of the genome, and hAT (Ac/Ds) elements alone represent 8%, more than in any other known plant genome. About 18% of the lotus genome is comprised of Copia LTR retrotransposons, and over 25% of them are associated with non-canonical termini (non-TGCA). Such high abundance of non-canonical LTR retrotransposons has not been reported for any other organism. TEs are very abundant in genic regions, with retrotransposons enriched in introns and DNA transposons primarily in flanking regions of genes. The recent insertion of TEs in introns has led to significant intron size expansion, with a total of 200 Mb in the 28 455 genes. This is accompanied by declining TE activity in intergenic regions, suggesting distinct control efficacy of TE amplification in different genomic compartments. Despite the prevalence of TEs in genic regions, some genes are associated with fewer TEs, such as those involved in fruit ripening and stress responses. Other genes are enriched with TEs, and genes in epigenetic pathways are the most associated with TEs in introns, indicating a dynamic interaction between TEs and the host surveillance machinery. The dramatic differential abundance of TEs with genes involved in different biological processes as well as the variation of target preference of different TEs suggests the composition and activity of TEs influence the path of evolution.
Keywords: Nelumbo nucifera; amplification; genes; intron; retrotransposon; target specificity; transposon.
© 2022 The Authors. The Plant Journal published by Society for Experimental Biology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures
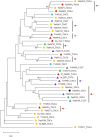
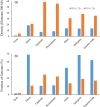
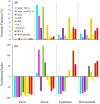
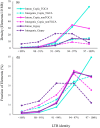

References
-
- Agren, J.A. & Wright, S.I. (2011) Co‐evolution between transposable elements and their hosts: a major factor in genome size evolution? Chromosome Research, 19, 777–786. - PubMed
-
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. & Lipman, D.J. (1990) Basic local alignment search tool. Journal of Molecular Biology, 215, 403–410. - PubMed
-
- Anderson, S.N. , Stitzer, M.C. , Brohammer, A.B. , Zhou, P. , Noshay, J.M. , O'Connor, C.H. et al. (2019) Transposable elements contribute to dynamic genome content in maize. The Plant Journal, 100, 1052–1065. - PubMed
-
- Badouin, H. , Gouzy, J. , Grassa, C.J. , Murat, F. , Staton, S.E. , Cottret, L. et al. (2017) The sunflower genome provides insights into oil metabolism, flowering and asterid evolution. Nature, 546, 148–152. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

