Loss of Nuclear Envelope Integrity and Increased Oxidant Production Cause DNA Damage in Adult Hearts Deficient in PKP2: A Molecular Substrate of ARVC
- PMID: 35959657
- PMCID: PMC9474627
- DOI: 10.1161/CIRCULATIONAHA.122.060454
Loss of Nuclear Envelope Integrity and Increased Oxidant Production Cause DNA Damage in Adult Hearts Deficient in PKP2: A Molecular Substrate of ARVC
Abstract
Background: Arrhythmogenic right ventricular cardiomyopathy (ARVC) is characterized by high propensity to life-threatening arrhythmias and progressive loss of heart muscle. More than 40% of reported genetic variants linked to ARVC reside in the PKP2 gene, which encodes the PKP2 protein (plakophilin-2).
Methods: We describe a comprehensive characterization of the ARVC molecular landscape as determined by high-resolution mass spectrometry, RNA sequencing, and transmission electron microscopy of right ventricular biopsy samples obtained from patients with ARVC with PKP2 mutations and left ventricular ejection fraction >45%. Samples from healthy relatives served as controls. The observations led to experimental work using multiple imaging and biochemical techniques in mice with a cardiac-specific deletion of Pkp2 studied at a time of preserved left ventricular ejection fraction and in human induced pluripotent stem cell-derived PKP2-deficient myocytes.
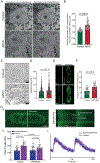
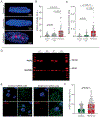
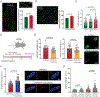
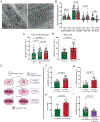
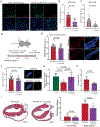
Results: Samples from patients with ARVC present a loss of nuclear envelope integrity, molecular signatures indicative of increased DNA damage, and a deficit in transcripts coding for proteins in the electron transport chain. Mice with a cardiac-specific deletion of Pkp2 also present a loss of nuclear envelope integrity, which leads to DNA damage and subsequent excess oxidant production (O2.- and H2O2), the latter increased further under mechanical stress (isoproterenol or exercise). Increased oxidant production and DNA damage is recapitulated in human induced pluripotent stem cell-derived PKP2-deficient myocytes. Furthermore, PKP2-deficient cells release H2O2 into the extracellular environment, causing DNA damage and increased oxidant production in neighboring myocytes in a paracrine manner. Treatment with honokiol increases SIRT3 (mitochondrial nicotinamide adenine dinucleotide-dependent protein deacetylase sirtuin-3) activity, reduces oxidant levels and DNA damage in vitro and in vivo, reduces collagen abundance in the right ventricular free wall, and has a protective effect on right ventricular function.
Conclusions: Loss of nuclear envelope integrity and subsequent DNA damage is a key substrate in the molecular pathology of ARVC. We show transcriptional downregulation of proteins of the electron transcript chain as an early event in the molecular pathophysiology of the disease (before loss of left ventricular ejection fraction <45%), which associates with increased oxidant production (O2.- and H2O2). We propose therapies that limit oxidant formation as a possible intervention to restrict DNA damage in ARVC.
Keywords: DNA damage; arrhythmogenic right ventricular dysplasia; nuclear envelope; oxidative stress; plakophilins; sirtuin 3.
Figures

Comment in
-
Top Stories: Mitochondrial origin of inherited cardiac arrhythmias.Heart Rhythm. 2024 Feb;21(2):235-236. doi: 10.1016/j.hrthm.2023.10.020. Heart Rhythm. 2024. PMID: 38296456 Free PMC article. No abstract available.
References
-
- Delmar M, McKenna WJ. The cardiac desmosome and arrhythmogenic cardiomyopathies: from gene to disease. Circ Res 2010;107, 700–714. - PubMed
-
- Corrado D, Link MS, Calkins H. Arrhythmogenic Right Ventricular Cardiomyopathy. N Engl J Med 2017;376, 61–72. - PubMed
-
- Groeneweg JA, Bhonsale, James CA, te Riele AS, Dooijes, Tichnell, Murray, Wiesfeld AC, Sawant AC, Kassamali et al. Clinical Presentation, Long-Term Follow-Up, and Outcomes of 1001 Arrhythmogenic Right Ventricular Dysplasia/Cardiomyopathy Patients and Family Members. Circ Cardiovasc Genet 2015;8, 437–446. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

