Closely related type II-C Cas9 orthologs recognize diverse PAMs
- PMID: 35959889
- PMCID: PMC9433092
- DOI: 10.7554/eLife.77825
Closely related type II-C Cas9 orthologs recognize diverse PAMs
Abstract
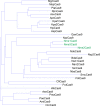
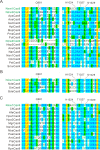
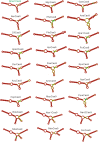
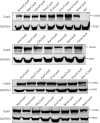
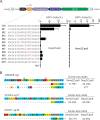
The RNA-guided CRISPR/Cas9 system is a powerful tool for genome editing, but its targeting scope is limited by the protospacer-adjacent motif (PAM). To expand the target scope, it is crucial to develop a CRISPR toolbox capable of recognizing multiple PAMs. Here, using a GFP-activation assay, we tested the activities of 29 type II-C orthologs closely related to Nme1Cas9, 25 of which are active in human cells. These orthologs recognize diverse PAMs with variable length and nucleotide preference, including purine-rich, pyrimidine-rich, and mixed purine and pyrimidine PAMs. We characterized in depth the activity and specificity of Nsp2Cas9. We also generated a chimeric Cas9 nuclease that recognizes a simple N4C PAM, representing the most relaxed PAM preference for compact Cas9s to date. These Cas9 nucleases significantly enhance our ability to perform allele-specific genome editing.
Keywords: CRISPR/Cas9; Nme1Cas9 orthologs; Nsp2Cas9; PAM diversity; crispr/cas9; genetics; genome editing; genomics; nme1cas9 orthologs; nsp2cas9; pam diversity.
© 2022, Wei et al.
Conflict of interest statement
JW, LH, JL, ZW, TQ, SG, SS No competing interests declared, SG, YW author on patent application number 202110878452X, "Cas9 protein, gene editing system containing Cas9 protein and application"; this patent relates to the technical field of gene editing, and specifically designs a CRISPR/Cas9 gene editing system and its application
Figures












References
-
- Agudelo D, Carter S, Velimirovic M, Duringer A, Rivest J-F, Levesque S, Loehr J, Mouchiroud M, Cyr D, Waters PJ, Laplante M, Moineau S, Goulet A, Doyon Y. Versatile and robust genome editing with streptococcus thermophilus CRISPR1-cas9. Genome Research. 2020;30:107–117. doi: 10.1101/gr.255414.119. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

