Noncoding RNAs in oral cancer
- PMID: 35959932
- PMCID: PMC10909450
- DOI: 10.1002/wrna.1754
Noncoding RNAs in oral cancer
Abstract
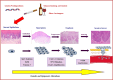
Oral cancer (OC) is the most prevalent subtype of cancer arising in the head and neck region. OC risk is mainly attributed to behavioral risk factors such as exposure to tobacco and excessive alcohol consumption, and a lesser extent to viral infections such as human papillomaviruses and Epstein-Barr viruses. In addition to these acquired risk factors, heritable genetic factors have shown to be associated with OC risk. Despite the high incidence, biomarkers for OC diagnosis are lacking and consequently, patients are often diagnosed in advanced stages. This delay in diagnosis is reflected by poor overall outcomes of OC patients, where 5-year overall survival is around 50%. Among the biomarkers proposed for cancer detection, noncoding RNA (ncRNA) can be considered as one of the most promising categories of biomarkers due to their role in virtually all cellular processes. Similar to other cancer types, changes in expressions of ncRNAs have been reported in OC and a number of ncRNAs have diagnostic, prognostic, and therapeutic potential. Moreover, some ncRNAs are capable of regulating gene expression by various mechanisms. Therefore, elucidating the current literature on the four main types of ncRNAs namely, microRNA, lncRNA, snoRNA, piwi-RNA, and circular RNA in the context of OC pathogenesis is timely and would enable further improvements and innovations in diagnosis, prognosis, and treatment of OC. This article is categorized under: RNA in Disease and Development > RNA in Disease RNA in Disease and Development > RNA in Development.
Keywords: diagnosis; noncoding RNA; oral cancer; prognosis; therapeutics.
© 2022 The Authors. WIREs RNA published by Wiley Periodicals LLC.
Conflict of interest statement
The authors have declared no conflicts of interest for this article.
Figures
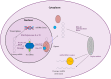


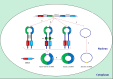
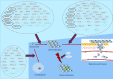
 , snoRNAs;
, snoRNAs;  , lncRNAs;
, lncRNAs;  , circular RNAs;
, circular RNAs;  , microRNAs.
, microRNAs.References
-
- American Society of Clinical Oncology (ASCO) . (2005. ‐2022). Cancer.net. https://www.cancer.net/cancer-types/oral-and-oropharyngeal-cancer/statis...
-
- Aghbari, S. M. H. , Gaafar, S. M. , Shaker, O. G. , Ashiry, S. E. , & Zayed, S. O. (2018). Evaluating the accuracy of microRNA27b and microRNA137 as biomarkers of activity and potential malignant transformation in oral lichen planus patients. Archives of Dermatological Research, 310(3), 209–220. 10.1007/s00403-018-1805-0 - DOI - PubMed
-
- Jain, A. , & Daaboul, H. E. (2019). Molecular pathogenesis of oral squamous cell carcinoma, squamous cell carcinoma ‐ Hallmark and treatment modalities. IntechOpen. 10.5772/intechopen.85650 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

