A gain-of-function allele of a DREB transcription factor gene ameliorates drought tolerance in wheat
- PMID: 35959993
- PMCID: PMC9614454
- DOI: 10.1093/plcell/koac248
A gain-of-function allele of a DREB transcription factor gene ameliorates drought tolerance in wheat
Abstract
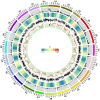
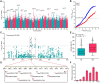
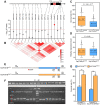
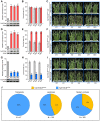
Drought is a major environmental factor limiting wheat production worldwide. However, the genetic components underlying wheat drought tolerance are largely unknown. Here, we identify a DREB transcription factor gene (TaDTG6-B) by genome-wide association study that is tightly associated with drought tolerance in wheat. Candidate gene association analysis revealed that a 26-bp deletion in the TaDTG6-B coding region induces a gain-of-function for TaDTG6-BDel574, which exhibits stronger transcriptional activation, protein interactions, and binding activity to dehydration-responsive elements (DRE)/CRT cis-elements than the TaDTG6-BIn574 encoded by the allele lacking the deletion, thus conferring greater drought tolerance in wheat seedlings harboring this variant. Knockdown of TaDTG6-BDel574 transcripts attenuated drought tolerance in transgenic wheat, whereas its overexpression resulted in enhanced drought tolerance without accompanying phenotypic abnormalities. Furthermore, the introgression of the TaDTG6-BDel574 elite allele into drought-sensitive cultivars improved their drought tolerance, thus providing a valuable genetic resource for wheat breeding. We also identified 268 putative target genes that are directly bound and transcriptionally regulated by TaDTG6-BDel574. Further analysis showed that TaDTG6-BDel574 positively regulates TaPIF1 transcription to enhance wheat drought tolerance. These results describe the genetic basis and accompanying mechanism driving phenotypic variation in wheat drought tolerance, and provide a novel genetic resource for crop breeding programs.
© American Society of Plant Biologists 2022. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures





Similar articles
-
TaDTGIP1-TaDTG6-BDel574-TaPIF1 module regulates drought stress response in wheat.New Phytol. 2025 Jun;246(5):2118-2136. doi: 10.1111/nph.70123. Epub 2025 Apr 7. New Phytol. 2025. PMID: 40195617
-
Variation in cis-regulation of a NAC transcription factor contributes to drought tolerance in wheat.Mol Plant. 2022 Feb 7;15(2):276-292. doi: 10.1016/j.molp.2021.11.007. Epub 2021 Nov 15. Mol Plant. 2022. PMID: 34793983
-
Transcription factor gene TaWRKY76 confers plants improved drought and salt tolerance through modulating stress defensive-associated processes in Triticum aestivum L.Plant Physiol Biochem. 2024 Nov;216:109147. doi: 10.1016/j.plaphy.2024.109147. Epub 2024 Sep 27. Plant Physiol Biochem. 2024. PMID: 39353294
-
Development of Drought-Tolerant Transgenic Wheat: Achievements and Limitations.Int J Mol Sci. 2019 Jul 8;20(13):3350. doi: 10.3390/ijms20133350. Int J Mol Sci. 2019. PMID: 31288392 Free PMC article. Review.
-
Transcription factors involved in drought tolerance and their possible role in developing drought tolerant cultivars with emphasis on wheat (Triticum aestivum L.).Theor Appl Genet. 2016 Nov;129(11):2019-2042. doi: 10.1007/s00122-016-2794-z. Epub 2016 Oct 13. Theor Appl Genet. 2016. PMID: 27738714 Review.
Cited by
-
Genome-wide identification and comprehensive analysis of the phytochrome-interacting factor (PIF) gene family in wheat.PLoS One. 2024 Jan 5;19(1):e0296269. doi: 10.1371/journal.pone.0296269. eCollection 2024. PLoS One. 2024. PMID: 38181015 Free PMC article.
-
The E3 ligase TaGW2 mediates transcription factor TaARR12 degradation to promote drought resistance in wheat.Plant Cell. 2024 Feb 26;36(3):605-625. doi: 10.1093/plcell/koad307. Plant Cell. 2024. PMID: 38079275 Free PMC article.
-
Transcriptional gene network involved in drought stress response: application for crop breeding in the context of climate change.Philos Trans R Soc Lond B Biol Sci. 2025 May 29;380(1927):20240236. doi: 10.1098/rstb.2024.0236. Epub 2025 May 29. Philos Trans R Soc Lond B Biol Sci. 2025. PMID: 40439309 Free PMC article. Review.
-
Genome-Wide Identification and Functional Analysis of the Genes of the ATL Family in Maize during High-Temperature Stress in Maize.Genes (Basel). 2024 Aug 22;15(8):1106. doi: 10.3390/genes15081106. Genes (Basel). 2024. PMID: 39202465 Free PMC article.
-
Transcriptome Analysis of Roots from Wheat (Triticum aestivum L.) Varieties in Response to Drought Stress.Int J Mol Sci. 2023 Apr 14;24(8):7245. doi: 10.3390/ijms24087245. Int J Mol Sci. 2023. PMID: 37108408 Free PMC article.
References
-
- Agarwal PK, Gupta K, Lopato S, Agarwal P (2017) Dehydration responsive element binding transcription factors and their applications for the engineering of stress tolerance. J Exp Bot 68: 2135–2148 - PubMed
-
- Ault TR (2020) On the essentials of drought in a changing climate. Science 368: 256–260 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

