Association Between Circulating CD4+ T Cell Methylation Signatures of Network-Oriented SOCS3 Gene and Hemodynamics in Patients Suffering Pulmonary Arterial Hypertension
- PMID: 35960497
- PMCID: PMC9944731
- DOI: 10.1007/s12265-022-10294-1
Association Between Circulating CD4+ T Cell Methylation Signatures of Network-Oriented SOCS3 Gene and Hemodynamics in Patients Suffering Pulmonary Arterial Hypertension
Abstract
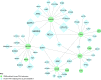
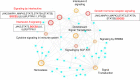
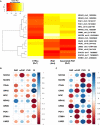
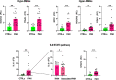
Pathogenic DNA methylation changes may be involved in pulmonary arterial hypertension (PAH) onset and its progression, but there is no data on potential associations with patient-derived hemodynamic parameters. The reduced representation bisulfite sequencing (RRBS) platform identified N = 631 differentially methylated CpG sites which annotated to N = 408 genes (DMGs) in circulating CD4+ T cells isolated from PAH patients vs. healthy controls (CTRLs). A promoter-restricted network analysis established the PAH subnetwork that included 5 hub DMGs (SOCS3, GNAS, ITGAL, NCOR2, NFIC) and 5 non-hub DMGs (NR4A2, GRM2, PGK1, STMN1, LIMS2). The functional analysis revealed that the SOCS3 gene was the most recurrent among the top ten significant pathways enriching the PAH subnetwork, including the growth hormone receptor and the interleukin-6 signaling. Correlation analysis showed that the promoter methylation levels of each network-oriented DMG were associated individually with hemodynamic parameters. In particular, SOCS3 hypomethylation was negatively associated with right atrial pressure (RAP) and positively associated with cardiac index (CI) (|r|≥ 0.6). A significant upregulation of the SOCS3, ITGAL, NFIC, NCOR2, and PGK1 mRNA levels (qRT-PCR) in peripheral blood mononuclear cells from PAH patients vs. CTRLs was found (P ≤ 0.05). By immunoblotting, a significant upregulation of the SOCS3 protein was confirmed in PAH patients vs. CTRLs (P < 0.01). This is the first network-oriented study which integrates circulating CD4+ T cell DNA methylation signatures, hemodynamic parameters, and validation experiments in PAH patients at first diagnosis or early follow-up. Our data suggests that SOCS3 gene might be involved in PAH pathogenesis and serve as potential prognostic biomarker.
Keywords: CD4+ T cells; DNA Methylation; Hemodynamic Parameters; Network Analysis; Pulmonary Arterial Hypertension.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- D'Alto M, Badagliacca R, Argiento P, Romeo E, Farro A, Papa S, Sarubbi B, Russo MG, Vizza CD, Golino P, Naeije R. Risk reduction and right heart reverse remodeling by upfront triple combination therapy in pulmonary arterial hypertension. Chest. 2020;157:376–383. doi: 10.1016/j.chest.2019.09.009. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

