The S-palmitoylome and DHHC-PAT interactome of Drosophila melanogaster S2R+ cells indicate a high degree of conservation to mammalian palmitoylomes
- PMID: 35960718
- PMCID: PMC9374236
- DOI: 10.1371/journal.pone.0261543
The S-palmitoylome and DHHC-PAT interactome of Drosophila melanogaster S2R+ cells indicate a high degree of conservation to mammalian palmitoylomes
Abstract
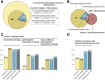
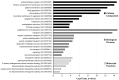
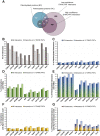
Protein S-palmitoylation, the addition of a long-chain fatty acid to target proteins, is among the most frequent reversible protein modifications in Metazoa, affecting subcellular protein localization, trafficking and protein-protein interactions. S-palmitoylated proteins are abundant in the neuronal system and are associated with neuronal diseases and cancer. Despite the importance of this post-translational modification, it has not been thoroughly studied in the model organism Drosophila melanogaster. Here we present the palmitoylome of Drosophila S2R+ cells, comprising 198 proteins, an estimated 3.5% of expressed genes in these cells. Comparison of orthologs between mammals and Drosophila suggests that S-palmitoylated proteins are more conserved between these distant phyla than non-S-palmitoylated proteins. To identify putative client proteins and interaction partners of the DHHC family of protein acyl-transferases (PATs) we established DHHC-BioID, a proximity biotinylation-based method. In S2R+ cells, ectopic expression of the DHHC-PAT dHip14-BioID in combination with Snap24 or an interaction-deficient Snap24-mutant as a negative control, resulted in biotinylation of Snap24 but not the Snap24-mutant. DHHC-BioID in S2R+ cells using 10 different DHHC-PATs as bait identified 520 putative DHHC-PAT interaction partners of which 48 were S-palmitoylated and are therefore putative DHHC-PAT client proteins. Comparison of putative client protein/DHHC-PAT combinations indicates that CG8314, CG5196, CG5880 and Patsas have a preference for transmembrane proteins, while S-palmitoylated proteins with the Hip14-interaction motif are most enriched by DHHC-BioID variants of approximated and dHip14. Finally, we show that BioID is active in larval and adult Drosophila and that dHip14-BioID rescues dHip14 mutant flies, indicating that DHHC-BioID is non-toxic. In summary we provide the first systematic analysis of a Drosophila palmitoylome. We show that DHHC-BioID is sensitive and specific enough to identify DHHC-PAT client proteins and provide DHHC-PAT assignment for ca. 25% of the S2R+ cell palmitoylome, providing a valuable resource. In addition, we establish DHHC-BioID as a useful concept for the identification of tissue-specific DHHC-PAT interactomes in Drosophila.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Neuronal palmitoyl acyl transferases exhibit distinct substrate specificity.FASEB J. 2009 Aug;23(8):2605-15. doi: 10.1096/fj.08-127399. Epub 2009 Mar 19. FASEB J. 2009. PMID: 19299482 Free PMC article.
-
Identification and Characterization of a Novel Palmitoyl Acyltransferase as a Druggable Rheostat of Dynamic Palmitoylome in L. donovani.Front Cell Infect Microbiol. 2018 Jun 20;8:186. doi: 10.3389/fcimb.2018.00186. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 29977865 Free PMC article.
-
Global identification of S-palmitoylated proteins and detection of palmitoylating (DHHC) enzymes in heart.J Mol Cell Cardiol. 2021 Jun;155:1-9. doi: 10.1016/j.yjmcc.2021.02.007. Epub 2021 Feb 23. J Mol Cell Cardiol. 2021. PMID: 33636221 Free PMC article.
-
Protein Palmitoylation by DHHC Protein Family.In: Kittler JT, Moss SJ, editors. The Dynamic Synapse: Molecular Methods in Ionotropic Receptor Biology. Boca Raton (FL): CRC Press/Taylor & Francis; 2006. Chapter 5. In: Kittler JT, Moss SJ, editors. The Dynamic Synapse: Molecular Methods in Ionotropic Receptor Biology. Boca Raton (FL): CRC Press/Taylor & Francis; 2006. Chapter 5. PMID: 21204476 Free Books & Documents. Review.
-
Emerging Roles of DHHC-mediated Protein S-palmitoylation in Physiological and Pathophysiological Context.Eur J Cell Biol. 2018 Jun;97(5):319-338. doi: 10.1016/j.ejcb.2018.03.005. Epub 2018 Mar 22. Eur J Cell Biol. 2018. PMID: 29602512 Review.
Cited by
-
Lost in traffic: consequences of altered palmitoylation in neurodegeneration.Front Physiol. 2023 May 30;14:1166125. doi: 10.3389/fphys.2023.1166125. eCollection 2023. Front Physiol. 2023. PMID: 37324388 Free PMC article. Review.
-
Recapitulating the potential contribution of protein S-palmitoylation in cancer.Cancer Metastasis Rev. 2024 Dec 27;44(1):20. doi: 10.1007/s10555-024-10217-3. Cancer Metastasis Rev. 2024. PMID: 39725785 Review.
References
-
- Sanders SS, Martin DDO, Butland SL, Lavallée-Adam M, Calzolari D, Kay C, et al.. Curation of the Mammalian Palmitoylome Indicates a Pivotal Role for Palmitoylation in Diseases and Disorders of the Nervous System and Cancers. Ben-Tal N, editor. PLoS Comput Biol. Public Library of Science; 2015;11: e1004405. doi: 10.1371/journal.pcbi.1004405 - DOI - PMC - PubMed
-
- Greaves J, Munro KR, Davidson SC, Riviere M, Wojno J, Smith TK, et al.. Molecular basis of fatty acid selectivity in the zDHHC family of S-acyltransferases revealed by click chemistry. Proc Natl Acad Sci USA. National Academy of Sciences; 2017;114: E1365–E1374. doi: 10.1073/pnas.1612254114 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

