The wound-activated ERF15 transcription factor drives Marchantia polymorpha regeneration by activating an oxylipin biosynthesis feedback loop
- PMID: 35960801
- PMCID: PMC9374346
- DOI: 10.1126/sciadv.abo7737
The wound-activated ERF15 transcription factor drives Marchantia polymorpha regeneration by activating an oxylipin biosynthesis feedback loop
Abstract
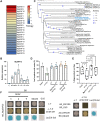
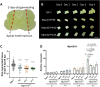
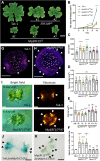
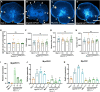
The regenerative potential in response to wounding varies widely among species. Within the plant lineage, the liverwort Marchantia polymorpha displays an extraordinary regeneration capacity. However, its molecular pathways controlling the initial regeneration response are unknown. Here, we demonstrate that the MpERF15 transcription factor gene is instantly activated after wounding and is essential for gemmaling regeneration following tissue incision. MpERF15 operates both upstream and downstream of the MpCOI1 oxylipin receptor by controlling the expression of oxylipin biosynthesis genes. The resulting rise in the oxylipin dinor-12-oxo-phytodienoic acid (dn-OPDA) levels results in an increase in gemma cell number and apical notch organogenesis, generating highly disorganized and compact thalli. Our data pinpoint MpERF15 as a key factor activating an oxylipin biosynthesis amplification loop after wounding, which eventually results in reactivation of cell division and regeneration. We suggest that the genetic networks controlling oxylipin biosynthesis in response to wounding might have been reshuffled over evolution.
Figures



Similar articles
-
Lipoxygenase pathway in model bryophytes: 12-oxo-9(13),15-phytodienoic acid is a predominant oxylipin in Physcomitrella patens.Phytochemistry. 2020 Dec;180:112533. doi: 10.1016/j.phytochem.2020.112533. Epub 2020 Oct 12. Phytochemistry. 2020. PMID: 33059187
-
An evolutionarily ancient fatty acid desaturase is required for the synthesis of hexadecatrienoic acid, which is the main source of the bioactive jasmonate in Marchantia polymorpha.New Phytol. 2022 Feb;233(3):1401-1413. doi: 10.1111/nph.17850. Epub 2021 Nov 30. New Phytol. 2022. PMID: 34846752
-
Dinor-12-oxo-phytodienoic acid conjugation with amino acids inhibits its phytohormone bioactivity in Marchantia polymorpha.Plant Physiol. 2024 Dec 23;197(1):kiae610. doi: 10.1093/plphys/kiae610. Plant Physiol. 2024. PMID: 39514772 Free PMC article.
-
MicroRNAs in Marchantia polymorpha.New Phytol. 2018 Oct;220(2):409-416. doi: 10.1111/nph.15294. Epub 2018 Jun 30. New Phytol. 2018. PMID: 29959894 Review.
-
Plant oxylipins: plant responses to 12-oxo-phytodienoic acid are governed by its specific structural and functional properties.FEBS J. 2009 Sep;276(17):4693-704. doi: 10.1111/j.1742-4658.2009.07195.x. Epub 2009 Aug 3. FEBS J. 2009. PMID: 19663904 Review.
Cited by
-
A blast from the past: Understanding stem cell specification in plant roots using laser ablation.Quant Plant Biol. 2023 Nov 28;4:e14. doi: 10.1017/qpb.2023.13. eCollection 2023. Quant Plant Biol. 2023. PMID: 38034417 Free PMC article.
-
Plant regeneration in the new era: from molecular mechanisms to biotechnology applications.Sci China Life Sci. 2024 Jul;67(7):1338-1367. doi: 10.1007/s11427-024-2581-2. Epub 2024 May 31. Sci China Life Sci. 2024. PMID: 38833085 Review.
-
Conservation of molecular responses upon viral infection in the non-vascular plant Marchantia polymorpha.Nat Commun. 2024 Sep 27;15(1):8326. doi: 10.1038/s41467-024-52610-0. Nat Commun. 2024. PMID: 39333479 Free PMC article.
-
Bridging the Gap: Genetic Insights into Graft Compatibility for Enhanced Kiwifruit Production.Int J Mol Sci. 2025 Mar 24;26(7):2925. doi: 10.3390/ijms26072925. Int J Mol Sci. 2025. PMID: 40243500 Free PMC article. Review.
References
-
- Heyman J., Canher B., Bisht A., Christiaens F., De Veylder L., Emerging role of the plant ERF transcription factors in coordinating wound defense responses and repair. J. Cell Sci. 131, jcs208215 (2018). - PubMed
-
- Ikeuchi M., Favero D. S., Sakamoto Y., Iwase A., Coleman D., Rymen B., Sugimoto K., Molecular mechanisms of plant regeneration. Annu. Rev. Plant Biol. 70, 377–406 (2019). - PubMed
-
- Ikeuchi M., Ogawa Y., Iwase A., Sugimoto K., Plant regeneration: Cellular origins and molecular mechanisms. Development 143, 1442–1451 (2016). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

