QM/MM Well-Tempered Metadynamics Study of the Mechanism of XBP1 mRNA Cleavage by Inositol Requiring Enzyme 1α RNase
- PMID: 35960929
- PMCID: PMC9472280
- DOI: 10.1021/acs.jcim.2c00735
QM/MM Well-Tempered Metadynamics Study of the Mechanism of XBP1 mRNA Cleavage by Inositol Requiring Enzyme 1α RNase
Abstract
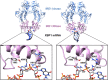
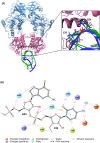
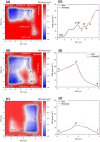
A range of in silico methodologies were herein employed to study the unconventional XBP1 mRNA cleavage mechanism performed by the unfolded protein response (UPR) mediator Inositol Requiring Enzyme 1α (IRE1). Using Protein-RNA molecular docking along with a series of extensive restrained/unrestrained atomistic molecular dynamics (MD) simulations, the dynamical behavior of the system was evaluated and a reliable model of the IRE1/XBP1 mRNA complex was constructed. From a series of well-converged quantum mechanics molecular mechanics well-tempered metadynamics (QM/MM WT-MetaD) simulations using the Grimme dispersion interaction corrected semiempirical parametrization method 6 level of theory (PM6-D3) and the AMBER14SB-OL3 force field, the free energy profile of the cleavage mechanism was determined, along with intermediates and transition state structures. The results show two distinct reaction paths based on general acid-general base type mechanisms, with different activation energies that perfectly match observations from experimental mutagenesis data. The study brings unique atomistic insights into the cleavage mechanism of XBP1 mRNA by IRE1 and clarifies the roles of the catalytic residues H910 and Y892. Increased understanding of the details in UPR signaling can assist in the development of new therapeutic agents for its modulation.
Conflict of interest statement
The authors declare the following competing financial interest(s): L.A.E. and E.C. are cofounders of Cell Stress Discoveries, Ltd. E.C. is cofounder of Thabor Therapeutics.
Figures

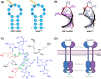

Similar articles
-
Herpes Simplex Virus 1 UL41 Protein Suppresses the IRE1/XBP1 Signal Pathway of the Unfolded Protein Response via Its RNase Activity.J Virol. 2017 Jan 31;91(4):e02056-16. doi: 10.1128/JVI.02056-16. Print 2017 Feb 15. J Virol. 2017. PMID: 27928013 Free PMC article.
-
Transcription of the NKG2D ligand MICA is suppressed by the IRE1/XBP1 pathway of the unfolded protein response through the regulation of E2F1.FASEB J. 2019 Mar;33(3):3481-3495. doi: 10.1096/fj.201801350RR. Epub 2018 Nov 19. FASEB J. 2019. PMID: 30452881
-
ER stress and distinct outputs of the IRE1α RNase control proliferation and senescence in response to oncogenic Ras.Proc Natl Acad Sci U S A. 2017 Sep 12;114(37):9900-9905. doi: 10.1073/pnas.1701757114. Epub 2017 Aug 28. Proc Natl Acad Sci U S A. 2017. PMID: 28847931 Free PMC article.
-
Getting RIDD of RNA: IRE1 in cell fate regulation.Trends Biochem Sci. 2014 May;39(5):245-54. doi: 10.1016/j.tibs.2014.02.008. Epub 2014 Mar 20. Trends Biochem Sci. 2014. PMID: 24657016 Review.
-
IRE1 signaling regulates chondrocyte apoptosis and death fate in the osteoarthritis.J Cell Physiol. 2022 Jan;237(1):118-127. doi: 10.1002/jcp.30537. Epub 2021 Jul 23. J Cell Physiol. 2022. PMID: 34297411 Free PMC article. Review.
Cited by
-
Novel Insights into the Catalytic Mechanism of Collagenolysis by Zn(II)-Dependent Matrix Metalloproteinase-1.Biochemistry. 2024 Aug 6;63(15):1925-1940. doi: 10.1021/acs.biochem.4c00076. Epub 2024 Jul 4. Biochemistry. 2024. PMID: 38963231 Free PMC article.
-
Multiscale In Silico Study of the Mechanism of Activation of the RtcB Ligase by the PTP1B Phosphatase.J Chem Inf Model. 2024 Feb 12;64(3):905-917. doi: 10.1021/acs.jcim.3c01600. Epub 2024 Jan 29. J Chem Inf Model. 2024. PMID: 38282538 Free PMC article.
References
-
- Almanza A.; Carlesso A.; Chintha C.; Creedican S.; Doultsinos D.; Leuzzi B.; Luis A.; McCarthy N.; Montibeller L.; More S.; Papaioannou A.; Puschel F.; Sassano M. L.; Skoko J.; Agostinis P.; de Belleroche J.; Eriksson L. A.; Fulda S.; Gorman A. M.; Healy S.; Kozlov A.; Munoz-Pinedo C.; Rehm M.; Chevet E.; Samali A. Endoplasmic Reticulum Stress Signalling–from Basic Mechanisms to Clinical Applications. FEBS journal 2019, 286, 241–278. 10.1111/febs.14608. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

