CHDbase: A Comprehensive Knowledgebase for Congenital Heart Disease-related Genes and Clinical Manifestations
- PMID: 35961607
- PMCID: PMC10372913
- DOI: 10.1016/j.gpb.2022.08.001
CHDbase: A Comprehensive Knowledgebase for Congenital Heart Disease-related Genes and Clinical Manifestations
Abstract
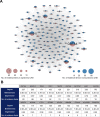
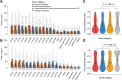
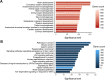
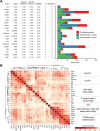
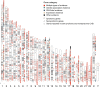
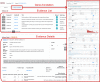
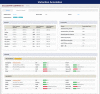
Congenital heart disease (CHD) is one of themost common causes of major birth defects, with a prevalence of 1%. Although an increasing number of studies have reported the etiology of CHD, the findings scattered throughout the literature are difficult to retrieve and utilize in research and clinical practice. We therefore developed CHDbase, an evidence-based knowledgebase of CHD-related genes and clinical manifestations manually curated from 1114 publications, linking 1124susceptibility genes and 3591 variations to more than 300 CHD types and related syndromes. Metadata such as the information of each publication and the selected population and samples, the strategy of studies, and the major findings of studies were integrated with each item of the research record. We also integrated functional annotations through parsing ∼ 50 databases/tools to facilitate the interpretation of these genes and variations in disease pathogenicity. We further prioritized the significance of these CHD-related genes with a gene interaction network approach and extracted a core CHD sub-network with 163 genes. The clear genetic landscape of CHD enables the phenotype classification based on the shared genetic origin. Overall, CHDbase provides a comprehensive and freely available resource to study CHD susceptibilities, supporting a wide range of users in the scientific and medical communities. CHDbase is accessible at http://chddb.fwgenetics.org.
Keywords: Classification; Congenital heart defect; Congenital heart disease; Database; Genetics.
Copyright © 2023 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors have declared no competing interests.
Figures



References
-
- Bernier P.L., Stefanescu A., Samoukovic G., Tchervenkov C.I. The challenge of congenital heart disease worldwide: epidemiologic and demographic facts. Semin Thorac Cardiovasc Surg Pediatr Card Surg Annu. 2010;13:26–34. - PubMed
-
- van der Linde D., Konings E.E., Slager M.A., Witsenburg M., Helbing W.A., Takkenberg J.J.M., et al. Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. J Am Coll Cardiol. 2011;58:2241–2247. - PubMed
-
- Oyen N., Poulsen G., Boyd H.A., Wohlfahrt J., Jensen P.K., Melbye M. Recurrence of congenital heart defects in families. Circulation. 2009;120:295–301. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

