Performance comparison of Agilent new SureSelect All Exon v8 probes with v7 probes for exome sequencing
- PMID: 35962321
- PMCID: PMC9375261
- DOI: 10.1186/s12864-022-08825-w
Performance comparison of Agilent new SureSelect All Exon v8 probes with v7 probes for exome sequencing
Abstract
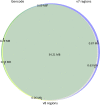
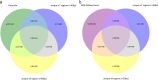
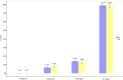
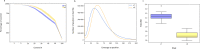
Exome sequencing is becoming a routine in health care, because it increases the chance of pinpointing the genetic cause of an individual patient's condition and thus making an accurate diagnosis. It is important for facilities providing genetic services to keep track of changes in the technology of exome capture in order to maximize throughput while reducing cost per sample. In this study, we focused on comparing the newly released exome probe set Agilent SureSelect Human All Exon v8 and the previous probe set v7. In preparation for higher throughput of exome sequencing using the DNBSEQ-G400, we evaluated target design, coverage statistics, and variants across these two different exome capture products. Although the target size of the v8 design has not changed much compared to the v7 design (35.24 Mb vs 35.8 Mb), the v8 probe design allows you to call more of SNVs (+ 3.06%) and indels (+ 8.49%) with the same number of raw reads per sample on the common target regions (34.84 Mb). Our results suggest that the new Agilent v8 probe set for exome sequencing yields better data quality than the current Agilent v7 set.
Keywords: Agilent SureSelect; BGI; Enrichment quality; Exome sequencing; MGISEQ; NGS; Variant calling; WES.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Comparative evaluation of four exome enrichment solutions in 2024: Agilent, Roche, Vazyme and Nanodigmbio.BMC Genomics. 2025 Jan 27;26(1):76. doi: 10.1186/s12864-024-11196-z. BMC Genomics. 2025. PMID: 39871131 Free PMC article.
-
Twist exome capture allows for lower average sequence coverage in clinical exome sequencing.Hum Genomics. 2023 May 3;17(1):39. doi: 10.1186/s40246-023-00485-5. Hum Genomics. 2023. PMID: 37138343 Free PMC article.
-
Comparison and evaluation of two exome capture kits and sequencing platforms for variant calling.BMC Genomics. 2015 Aug 5;16(1):581. doi: 10.1186/s12864-015-1796-6. BMC Genomics. 2015. PMID: 26242175 Free PMC article.
-
Performance comparison of four commercial human whole-exome capture platforms.Sci Rep. 2015 Aug 3;5:12742. doi: 10.1038/srep12742. Sci Rep. 2015. PMID: 26235669 Free PMC article.
-
Whole Exome Library Construction for Next Generation Sequencing.Methods Mol Biol. 2018;1706:163-174. doi: 10.1007/978-1-4939-7471-9_9. Methods Mol Biol. 2018. PMID: 29423798 Review.
Cited by
-
Comparative evaluation of four exome enrichment solutions in 2024: Agilent, Roche, Vazyme and Nanodigmbio.BMC Genomics. 2025 Jan 27;26(1):76. doi: 10.1186/s12864-024-11196-z. BMC Genomics. 2025. PMID: 39871131 Free PMC article.
-
Twist exome capture allows for lower average sequence coverage in clinical exome sequencing.Hum Genomics. 2023 May 3;17(1):39. doi: 10.1186/s40246-023-00485-5. Hum Genomics. 2023. PMID: 37138343 Free PMC article.
-
Bioinformatics of germline variant discovery for rare disease diagnostics: current approaches and remaining challenges.Brief Bioinform. 2024 Jan 22;25(2):bbad508. doi: 10.1093/bib/bbad508. Brief Bioinform. 2024. PMID: 38271481 Free PMC article. Review.
-
Comparative analysis of whole exome sequencing kits for the canine genome.PLoS One. 2024 Nov 4;19(11):e0312203. doi: 10.1371/journal.pone.0312203. eCollection 2024. PLoS One. 2024. PMID: 39495750 Free PMC article.
References
MeSH terms
Grants and funding
- 075-15-2019-1789/Ministry of Science and Higher Education of the Russian Federation
- 075-15-2019-1789/Ministry of Science and Higher Education of the Russian Federation
- 075-15-2019-1789/Ministry of Science and Higher Education of the Russian Federation
- 075-15-2019-1789/Ministry of Science and Higher Education of the Russian Federation
- 075-15-2019-1789/Ministry of Science and Higher Education of the Russian Federation
- 075-15-2019-1789/Ministry of Science and Higher Education of the Russian Federation
- 075-15-2019-1789/Ministry of Science and Higher Education of the Russian Federation
- 075-15-2019-1789/Ministry of Science and Higher Education of the Russian Federation
- 075-15-2019-1789/Ministry of Science and Higher Education of the Russian Federation
LinkOut - more resources
Full Text Sources
Miscellaneous

