Genomic analysis reveals the role of integrative and conjugative elements in plant pathogenic bacteria
- PMID: 35962419
- PMCID: PMC9373382
- DOI: 10.1186/s13100-022-00275-1
Genomic analysis reveals the role of integrative and conjugative elements in plant pathogenic bacteria
Abstract
Background: ICEs are mobile genetic elements found integrated into bacterial chromosomes that can excise and be transferred to a new cell. They play an important role in horizontal gene transmission and carry accessory genes that may provide interesting phenotypes for the bacteria. Here, we seek to research the presence and the role of ICEs in 300 genomes of phytopathogenic bacteria with the greatest scientific and economic impact.
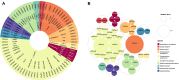
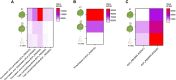
Results: Seventy-eight ICEs (45 distinct elements) were identified and characterized in chromosomes of Agrobacterium tumefaciens, Dickeya dadantii, and D. solani, Pectobacterium carotovorum and P. atrosepticum, Pseudomonas syringae, Ralstonia solanacearum Species Complex, and Xanthomonas campestris. Intriguingly, the co-occurrence of four ICEs was observed in some P. syringae strains. Moreover, we identified 31 novel elements, carrying 396 accessory genes with potential influence on virulence and fitness, such as genes coding for functions related to T3SS, cell wall degradation and resistance to heavy metals. We also present the analysis of previously reported data on the expression of cargo genes related to the virulence of P. atrosepticum ICEs, which evidences the role of these genes in the infection process of tobacco plants.
Conclusions: Altogether, this paper has highlighted the potential of ICEs to affect the pathogenicity and lifestyle of these phytopathogens and direct the spread of significant putative virulence genes in phytopathogenic bacteria.
Keywords: Genome evolution; Horizontal gene transfer; Mobile genetic elements (MGE); Phytopathology.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Guglielmini, J., Quintais, L., Garcillán-Barcia, M. P., de la Cruz, F., & Rocha, E. P. C. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genetics. 2011;7(8). 10.1371/journal.pgen.1002222. 78(1), 138–157. 10.1111/j.1365-2958.2010.07317.x - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

