The volume changes of unfolding of dsDNA
- PMID: 35962547
- PMCID: PMC9811605
- DOI: 10.1016/j.bpj.2022.08.005
The volume changes of unfolding of dsDNA
Abstract
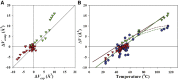
High hydrostatic pressure can have profound effects on the stability of biomacromolecules. The magnitude and direction (stabilizing or destabilizing) of this effect is defined by the volume changes in the system, ΔV. Positive volume changes will stabilize the starting native state, whereas negative volume changes will lead to the stabilization of the final unfolded state. For the DNA double helix, experimental data suggested that when the thermostability of dsDNA is below 50°C, increase in hydrostatic pressure will lead to destabilization; i.e., helix-to-coil transition has negative ΔV. In contrast, the dsDNA sequences with the thermostability above 50°C showed positive ΔV values and were stabilized by hydrostatic pressure. In order to get insight into this switch in the response of dsDNA to hydrostatic pressure as a function of temperature, first we further validated this trend using experimental measurements of ΔV for 10 different dsDNA sequences using pressure perturbation calorimetry. We also developed a computational protocol to calculate the expected volume changes of dsDNA unfolding, which was benchmarked against the experimental set of 50 ΔV values that included, in addition to our data, the values from the literature. Computation predicts well the experimental values of ΔV. Such agreement between computation and experiment lends credibility to the computation protocol and provides molecular level rational for the observed temperature dependence of ΔV that can be traced to the hydration. Difference in the ΔV value for A/T versus G/C basepairs is also discussed.
Copyright © 2022 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interest.
Figures




References
-
- Cech T.R. Structural biology. The ribosome is a ribozyme. Science. 2000;289:878–879. - PubMed
-
- Wochner A., Attwater J., et al. Holliger P. Ribozyme-catalyzed transcription of an active ribozyme. Science. 2011;332:209–212. - PubMed
-
- Zaug A.J., Cech T.R. The Tetrahymena intervening sequence ribonucleic acid enzyme is a phosphotransferase and an acid phosphatase. Biochemistry. 1986;25:4478–4482. - PubMed
-
- Condie K.C. Academic Press; 2021. Earth as an Evolving Planetary System.
-
- Gilbert W. Origin of life: the RNA world. Nature. 1986;319:618.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

