Autopolyploid inheritance and a heterozygous reciprocal translocation shape chromosome genetic behavior in tetraploid blueberry (Vaccinium corymbosum)
- PMID: 35962608
- PMCID: PMC10087351
- DOI: 10.1111/nph.18428
Autopolyploid inheritance and a heterozygous reciprocal translocation shape chromosome genetic behavior in tetraploid blueberry (Vaccinium corymbosum)
Abstract
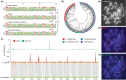
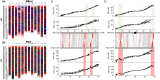
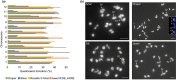
Understanding chromosome recombination behavior in polyploidy species is key to advancing genetic discoveries. In blueberry, a tetraploid species, the line of evidences about its genetic behavior still remain poorly understood, owing to the inter-specific, and inter-ploidy admixture of its genome and lack of in depth genome-wide inheritance and comparative structural studies. Here we describe a new high-quality, phased, chromosome-scale genome of a diploid blueberry, clone W85. The genome was integrated with cytogenetics and high-density, genetic maps representing six tetraploid blueberry cultivars, harboring different levels of wild genome admixture, to uncover recombination behavior and structural genome divergence across tetraploid and wild diploid species. Analysis of chromosome inheritance and pairing demonstrated that tetraploid blueberry behaves as an autotetraploid with tetrasomic inheritance. Comparative analysis demonstrated the presence of a reciprocal, heterozygous, translocation spanning one homolog of chr-6 and one of chr-10 in the cultivar Draper. The translocation affects pairing and recombination of chromosomes 6 and 10. Besides the translocation detected in Draper, no other structural genomic divergences were detected across tetraploid cultivars and highly inter-crossable wild diploid species. These findings and resources will facilitate new genetic and comparative genomic studies in Vaccinium and the development of genomic assisted selection strategy for this crop.
Keywords: autopolyploid; blueberry (Vaccinium corymbosum L.); centromeric repeat; chromosomal translocation; chromosome structure; phased genome; polyploid genetic behavior.
© 2022 The Authors. New Phytologist © 2022 New Phytologist Foundation.
Conflict of interest statement
None declared.
Figures



References
-
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B (Methodological) 57: 289–300.
-
- Bourke PM, Arens P, Voorrips RE, Esselink GD, Koning‐Boucoiran CFS, van't Westende WPC, Santos Leonardo T, Wissink P, Zheng C, van Geest G et al. 2017. Partial preferential chromosome pairing is genotype dependent in tetraploid rose. The Plant Journal 90: 330–343. - PubMed
-
- Brevis PA, Bassil NV, Ballington JR, Hancock JF. 2008. Impact of wide hybridization on highbush blueberry breeding. Journal of the American Society for Horticultural Science 133: 427–437.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

